Chapter: Genetics and Molecular Biology: Induction, Repression, and the araBAD Operon
Binding Sites of the ara Regulatory Proteins
Binding Sites of the ara Regulatory Proteins
Now that
the regulatory phenomena of the ara
operon have been laid out, what can be said about its mechanism of regulation?
The nucleotide sequences of the promoters pBAD,
pE, pFGH, show three distinct regions of homology among
themselves. One of these is the RNA polymerase-binding site. Another is the
cyclic AMP receptor protein-binding site, and the third is the AraC
protein-binding site. Inexplicably, the araBAD
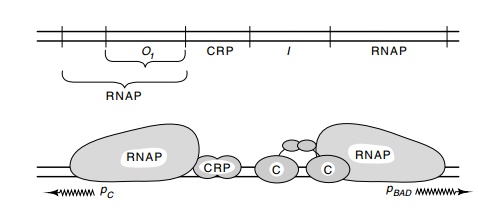
Figure
12.10 Schematic of the binding sites of
AraC protein, CRP, and RNApolymerase on the araBAD
regulatory region. The araO1 site is the operator for pCand the araI site is required for induction.
RNA
polymerase-binding site is quite similar to the RNA polymerase sites of
promoters that do not require auxiliary proteins for activity. Since pBAD requires the auxiliary
proteins AraC and CRP for its activity, the polymerase-binding sequence might
have been expected to be mark-edly different from the consensus RNA
polymerase-binding sequence.
The
identity of the protein-binding sites was established by DNAse footprinting. In
araBAD, the AraC protein-binding site
is just upstream from RNA polymerase, and the CRP-binding site is just upstream
from that of araC (Fig. 12.10). The
AraC-binding site is called araI for
induction. These three sites are required for induction of pBAD.
A second
AraC protein-binding site called araO1
lies another 60 nucleotides upstream of araI.
This site overlaps the RNA polymerase-binding site of pC, and since occupancy of araO1 by AraC protein sterically interferes with the
binding of RNA polymerase at pC,
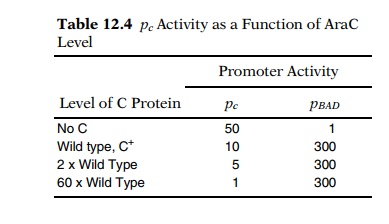
araO1 is an operator of pC. AraO1 is not directly involved with repression of pBAD. A simple demonstration
of this is the behavior of pC
and pBAD as the level of
AraC protein is increased in a series of strains containing plasmids with araC fused to promoters of different
strengths. As the level of AraC
Deletion
analysis showed that one of the sites required for repression of pBAD lies beyond araO1. It is called araO2, and it lies 270 base
pairs upstream from the pBAD
transcription start point. Footprinting has confirmed that AraC protein binds
to this site, and mutation analysis
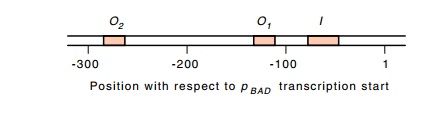
has shown
that a single nucleotide change in this region can eliminate repression of pBAD. Deletion of this site
is the reason that the original deletion that was isolated by Englesberg could
no longer repress pBAD but
could induce. His deletion extended through araO2
into araO1, and during the
interval of several years between the discovery of O1 and the discovery of O2, araO1
was thought to be required for repression of pBAD.
The
location of the CRP site required for stimulation of the pBAD promoter is most surprising. In the lac operon, the CRP site lies at
positions -48 to -78 with respect to the start of transcription, just next to
RNA polymerase. At the ara promoter,
AraC protein occupies this site and CRP lies further away, binding to positions
-80 to -110. CRP makes specific contacts with RNA polymerase in activating
transcription of the lac promoter.
Does it make a different set of specific contacts with AraCprotein, and does
AraC protein make yet another set of contacts with RNA polymerase?
Alternatively, both proteins might simultaneously contact RNA polymerase to
activate transcription, but it seems more likely that CRP activates through a
DNA bending mechanism and that AraC activates through direct protein-protein
contacts.
Related Topics