Chapter: Genetics and Molecular Biology: Induction, Repression, and the araBAD Operon
How AraC Protein Loops and Unloops
How AraC Protein Loops and Unloops
Two lines of in vivo evidence suggest that the loop between araI and araO2is broken upon the addition of arabinose. First, the operon is fullyinducible if araO2 is deleted. This implies that the loop is not involved with induction. Second, the occupancy of araO2 falls immediately after arabinose addition. One good way to study such regulated looping is to perform in vitro experiments. AraC loops sufficiently weakly, however, that looping of linear DNA just does not occur. Instead, looping had to be studied using small supercoiled circles of about 400 base pairs. These are sufficiently small, and the supercoiled DNA is sufficiently wound upon itself, that AraC protein can easily bridge the gap between araO2 and araI. These supercoiled, looped circles migrate upon electrophoresis at a rate different from free circles or circles with AraC bound at a single point. Therefore binding, looping, and unlooping can all be assayed.
Study of
the looped supercoils revealed an unexpected property. As AraC is a dimer in
solution, and a dimer binds to linear DNA containing the araI site, it had seemed likely that the looped species would be
formed from a dimer of AraC bound to araI
and a dimer bound to araO2.
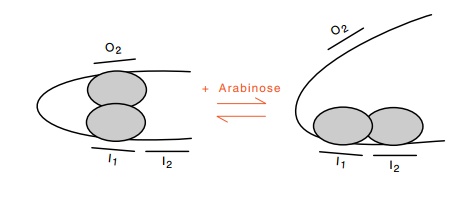
This proved not to be the case. The looped species contains only one dimer! In
the absence of arabinose one of the monomers of an AraC dimer binds to the left
half of araI, and the other monomer
binds to araO2(Fig.
12.13). The left and right halves of araI
are called araI1 and araI2.
Upon the addition of arabinose the protein reorients, and thesubunit contacting
araO2 lets go, and
contacts araI2. The
subunit rear-rangement occurs in the absence of free protein and is largely
inde-pendent of the precise sequences at the sites involved. In the absence of
arabinose the protein prefers to loop, that is, contact to nonlocal sites. In
the presence of arabinose, the protein prefers to contact local sites. The
contacting of the araI2
site by AraC protein provides the inducing signal to RNA polymerase. Only if
this site is properly positioned so as to overlap partially the -35 sequence of
pBAD does AraC activate
transcription.
Figure
12.13 Reorientation of the two subunits
of AraC protein shifts the mostfavored conformation from a looped state to the
unlooped state.
One
simple mechanism that could generate the observed behavior of AraC protein is a
subunit reorientation. Suppose that in the absence of arabinose the subunits
are oriented such that binding to two half-sites in a looping structure is
energetically easier than binding to two adja-cent half-sites. Then the protein
would prefer to loop in the absence of arabinose. If the presence of arabinose
causes the subunits to reorient so that binding to the two half-sites of araI is favored, then the protein would
unloop, bind to both halves of araI,
and induce pBAD.
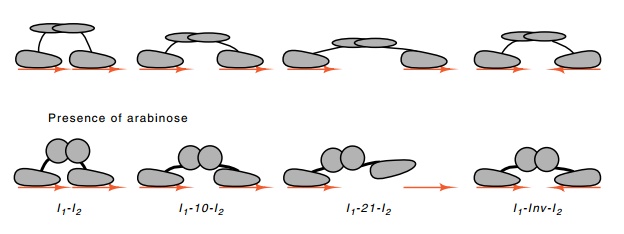
Figure 12.14 Binding of AraC protein to sequences with different distancesseparating the half-sites or with one half-site inverted. In the presence of arabinose, the reach of AraC is reduced.
The data
supports a slight modification of the reorientation model outlined above. AraC
protein monomers consist of a dimerization domain flexibly connected to a
DNA-binding domain. The half-sites to which AraC binds normally are oriented in
a direct repeat orientation. AraC still binds with high affinity when one
half-site is inverted so that the total site possesses inverted repeat
symmetry. This shows that the dimerization and DNA-binding domains are flexibly
connected. Fur-ther, the half-sites of araI
can be separated by an additional 10 or 21 base pairs, or even inverted,
without greatly affecting the affinity of AraC. When, however, arabinose is
added, the affinity of AraC for the wild-type araI site, the “+10” site, and the inverted site is increased, but
that for the “+21” site is decreased. Because arabinose did not increase the
affinity of AraC for all the sites, arabinose cannot be increasing the
intrinsic affinity of a DNA-binding domain for a half-site. Instead, arabinose
alters the relative positions of the DNA-binding domains or changes the
flexibility or length of the connection between the dimeri - zation and
DNA-binding domains (Fig. 12.14).
Related Topics