Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Bacterial Processes
Regulation and Adaptation - Cell Growth of Bacterial
REGULATION AND ADAPTATION
Metabolic reactions must proceed in a coordinated fashion. It would not do to have them governed solely by the laws of “mass action” by which the concentrations of reactants and products determine the rate of reactions. Furthermore, it would not do to have rates of individual reactions set at some fixed levels. Bacteria can do little to control their environ-ment, and any change in environment (eg, in temperature, pH, nutrient availability, osmo-larity) would disrupt any preset synchronization or render it inappropriate. Bacteria must, therefore, not just coordinate reactions, but must do so in a flexible, adjustable manner to make growth possible in a changing environment. They accomplish this feat by many reg-ulatory mechanisms, some of which operate to control enzyme activity, some to control gene expression.
Control of Enzyme Activity
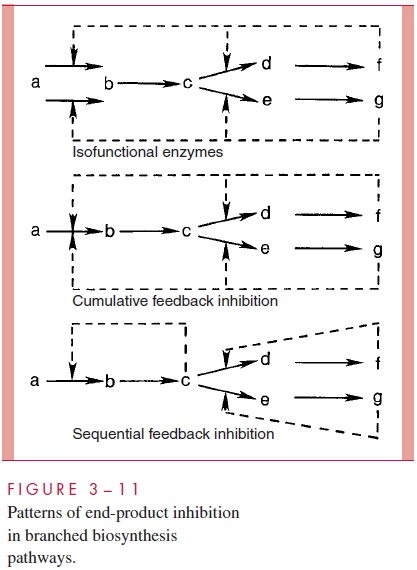
Although there are many examples of covalent modification of enzymes (eg, by phospho-rylation, methylation, or acylation) to alter their activity, by far the most prevalent means by which bacterial cells modulate the flow of material through fueling and biosynthetic pathways is by changing the activity of allosteric enzymes through the reversible binding of low-molecular-weight metabolites (ligands). In fueling pathways it is common for AMP, ADP, and ATP to control the activity of enzymes by causing conformational changes of allosteric enzymes, usually located at critical branch points where pathways intersect. By this means, the flow of carbon from the major substrates through the various pathways is adjusted to be appropriate to the demands of biosynthesis. For example, the energy charge of the cell, defined as (ATP + 1 ⁄ 2 ADP)/(ATP+ADP+AMP), is keptvery close to 0.85 under all conditions of growth and nongrowth. In biosynthetic path-ways, it is common for the end product of the pathway to control the activity of the first enzyme in the pathway. This pattern, called feedback inhibition or end-product inhibi-tion, ensures that each building block is made at exactly the rate it is being used for po-lymerization. It also ensures that building blocks supplied in the medium are not waste-fully duplicated by synthesis. Because many biosynthetic pathways are branched and have multiple end products, special arrangements must be made to produce effective reg-ulation. These include the production of multiple isofunctional enzymes for the controlled step, the design of allosteric enzymes that require the cumulative effect of all end prod-ucts to be completely inhibited, and sequential inhibition of each subpathway by its last product (Fig 3 – 11).
Control of Gene Expression
To a far greater extent than eukaryotic cells, bacteria regulate their metabolism by chang-ing the amounts of different enzymes. This is accomplished chiefly by governing their rates of synthesis, that is, by controlling gene expression. This works rapidly for bacteria because of their speed of growth; shutting off the synthesis of a particular enzyme results in short order in the reduction of its cellular level due to dilution by the growth of the cell. Most importantly, bacterial mRNA is degraded rapidly. With an average half-life of 2 to 3 minutes at 37°C, the mRNA complement of the cell can be totally changed in a small fraction of a generation time. The synthesis of a given enzyme can therefore be rapidly turned on and just as rapidly turned off simply by changes in the rate of transcription of its gene.
Most, although not all, of the regulation of gene expression occurs at or near the beginning of the process: the initiation of transcription. That is, gene expression is not regulated by changing the rate of mRNA chain elongation; once started, transcription pro-ceeds at a more or less constant rate. Regulation occurs by a decision of whether to initi-ate or not, or what amounts to the same thing, by setting the frequency of initiation.
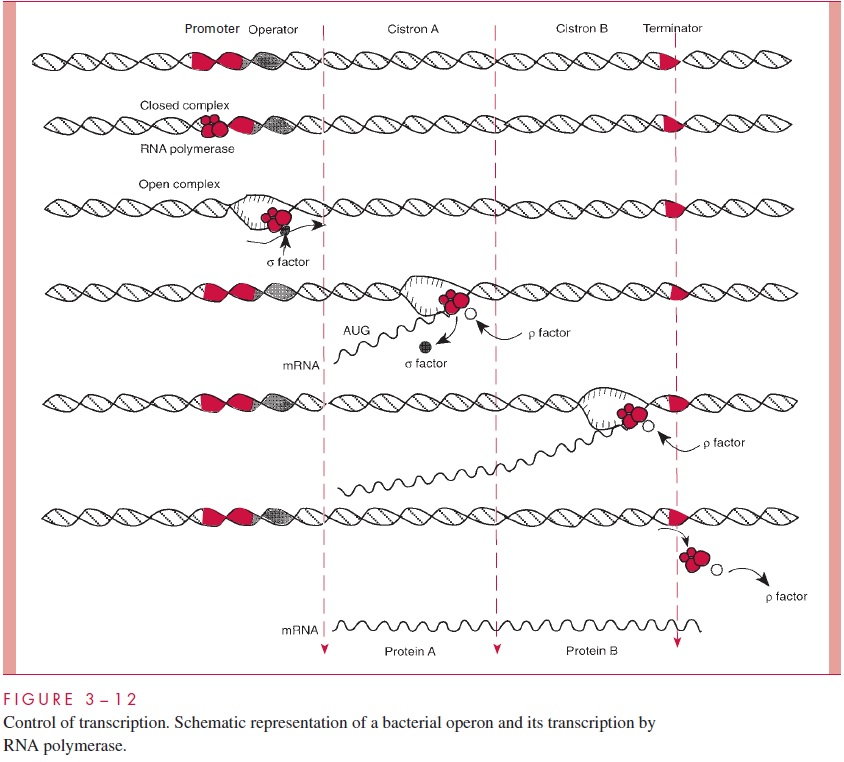
A closer look at transcription is necessary to understand how it is controlled. Most of the genes we know about in bacteria are organized asmulticistronic operons. A cistron is a segment of DNA encoding a polypeptide. An operon is the unit of transcription; the cistrons that it comprises are cotranscribed as a single mRNA. The structure of a typical operon (Fig 3 – 12) consists of a promoter region, an operator region, component cistrons, and a terminator. In the best-studied bacterium, E. coli, RNA polymerase, pro-grammed by the major replaceable subunit, σ-70, recognizes the promoter region and binds to the DNA. Initially the binding is a closed complex, but this can be converted into an open complex in which the two strands of DNA are partially separated. Strand separa-tion exposes the nucleotide bases and permits initiation of synthesis of a mRNA strand complementary to the sense strand of the DNA. In a simple case, transcription continues through the cistrons of the operon until the termination signal is reached. In some cases recognition of the termination signal requires another removable subunit of RNA poly-merase, ρ. This process is shown in Figure 3 – 12.
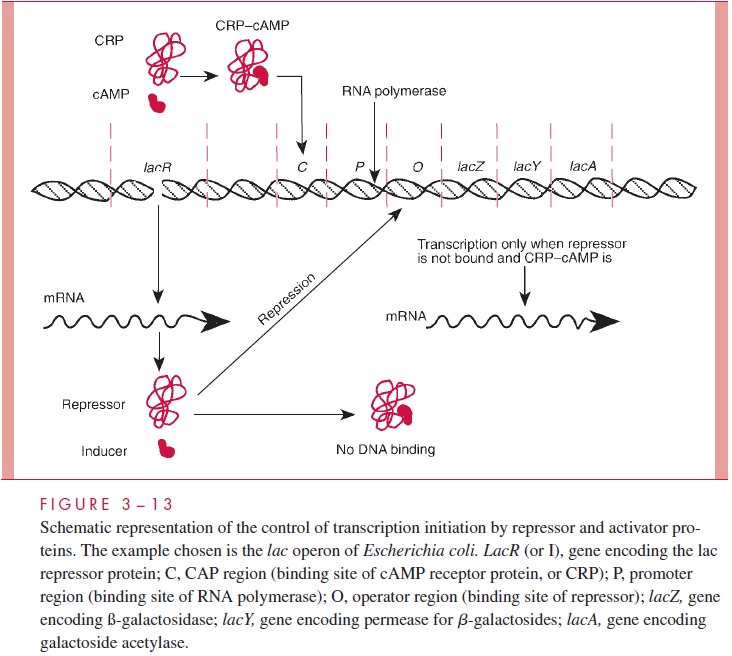
Near the promoter in many operons is an operator to which a specific regulator pro-tein or transcription factor can bind. In some cases the binding of this regulator blocksinitiation; in such a case of negative control, the regulator is called a repressor. Repres-sors are allosteric proteins, and their binding to the operator depends on their conforma-tion, which is determined by the binding of ligands that are called corepressors if their action permits binding of the repressor and inducers if their action prevents binding. In some cases, the regulator protein is required for initiation of transcription, and it is then called an activator. The functioning of both types of regulator proteins on transcription initiation is illustrated in Figure 3 – 13 using the regulation of the lac operon as an exam-ple. This operon encodes proteins necessary for the use of lactose as a carbon and energy source.
Some regulator proteins bend DNA on binding, and this can bring together what would otherwise be distant sites of the DNA. In this manner, proteins bound at sites called enhancers far upstream or downstream of a promoter can be brought into physical contact with RNA polymerase and influence its activity. One such DNA bender in E. coli is called the integration host factor.
Many regulator proteins are converted from inactive to active forms by covalent mod-ification rather than by the allosteric binding of a ligand. Phosphorylation is by far the most common modifying event and operates in the widespread two-component signaltransduction pathways described below under cell stress regulons.
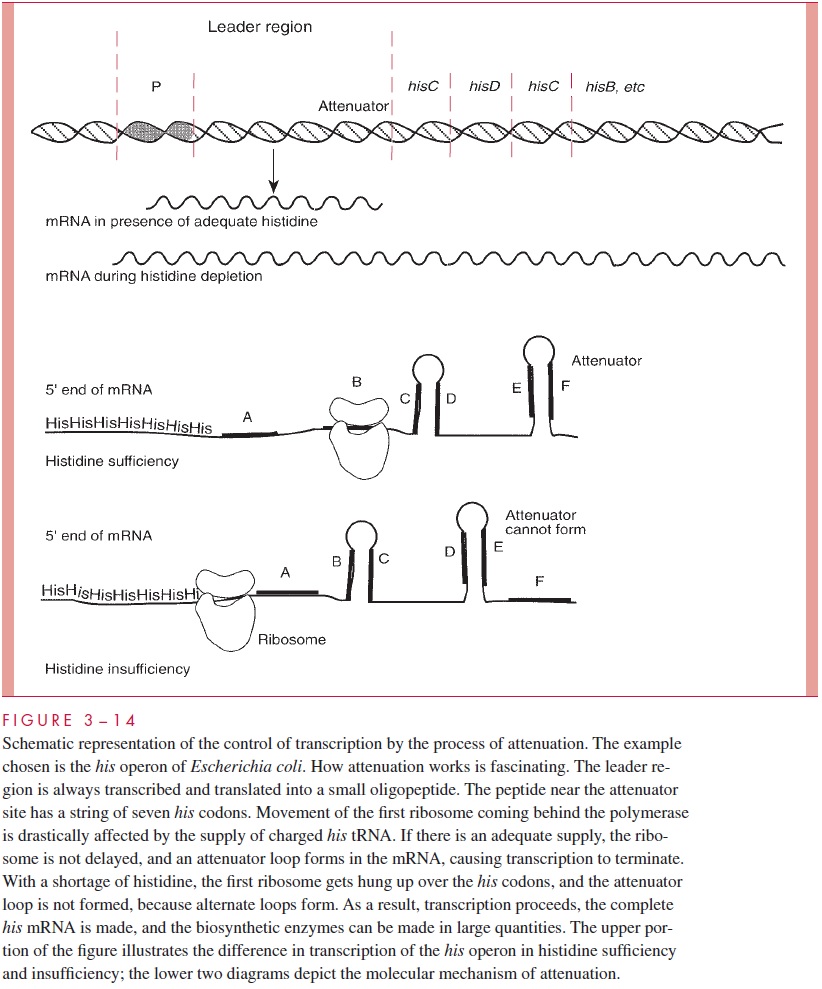
Once transcription is initiated it may continue uneventfully, but in some operons an-other site of control is quickly encountered. After transcription of a leader region, the RNA polymerase encounters a region known as an attenuator. Synthesis of mRNA is aborted at the attenuator; only a small percentage of the RNA polymerase molecules reaching the attenuator can successfully pass through it. However, the activity of the at-tenuator can be modified by a process that involves not a regulator protein but rather changes in the secondary structure of the mRNA. This regulatory process is illustrated in Figure 3 – 14 using the his operon, which encodes the enzymes necessary for the biosyn-thesis of the amino acid L-histidine, as an example. In enteric bacteria, attenuation is a common means of controlling biosynthetic operons. Note that it differs from the repres-sion mechanism in that it requires no special regulatory gene or regulatory proteins.
There are many instances known in which groups of genes that are independently controlled as members of different operons must cooperate to accomplish some response to an environmental change. When such a group of genes is subject to the control of a common regulator, the group is called a regulon. One such regulon, or global controlsystem, is catabolite repression. Its function is to prevent the cell from responding to thepresence of alternative carbon sources when the environment already provides a more than adequate supply from the preferred substrate, glucose. This control is brought about as follows. Operons that encode catabolic enzymes (those responsible for initiating the use of carbon sources, such as lactose, maltose, arabinose, and other sugars and amino acids) have weak promoters that need help to promote high-level initiation of transcrip-tion by RNA polymerase.
The help is supplied by a regulator protein called cataboliteactivator protein or cAMP receptor protein (CRP). This protein, if and only if cyclicAMP is bound to it, binds slightly upstream from the promoter and permits high-level ex-pression if the operon is specifically induced (and the repressor has been removed by in-duction). Because cAMP levels are very low during growth on glucose or other favored substrates, there is insufficient cAMP – CRP complex to activate catabolic operons even if their inducers are present in the environment. As a result, the cells ignore the induction signal if they have an adequate supply of glucose.
Finally, gene regulation in bacteria is accomplished by unique tactics so far discov-ered only in pathogens. These are included in the following section.
Related Topics