Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Bacterial Processes
Polymerization Reactions - Cell Growth of Bacterial
Polymerization Reactions
Unlike fueling and biosynthetic processes, polymerization reactions offer many targets for antimicrobic chemotherapy. The reason is simple: the bacterial machineries for replication, transcription, and translation differ from that in the human host cells.
Polymerization of DNA is called replication. From studies largely made inE. coli,DNA replication involves 12 or more proteins acting at a small number of sites (replica-tion forks) where DNA is synthesized from activated building blocks (dATP, dGTP, dCTP, and TTP). Replication always begins at special sites on the chromosome called oriC in E. coli (for origin of replication) and then proceeds bidirectionally around the cir-cular chromosome (Fig 3 – 4). Synthesis of DNA at each replication fork is termed semi-conservative because each of the DNA chains serves as the template for the synthesis ofits complement, and, therefore, one of the two chains of the new double-stranded mole-cule is conserved from the original chromosome. One of the two new strands must be synthesized in chemically the opposite direction of the other; this is accomplished by hav-ing each new strand made in short segments, 5’ to 3’ , which are then ligated by one of the DNA-synthesizing enzymes (see Fig 3 – 4). Interestingly, an RNA primer is involved in getting each of these segments initiated. The two replication forks meet at the opposite side of the circle. The frequency of initiation of chromosome replication (and, therefore, the number of growing points) varies with cell growth rate; the chain elongation rate is rather constant at a given temperature independent of cell growth rate.
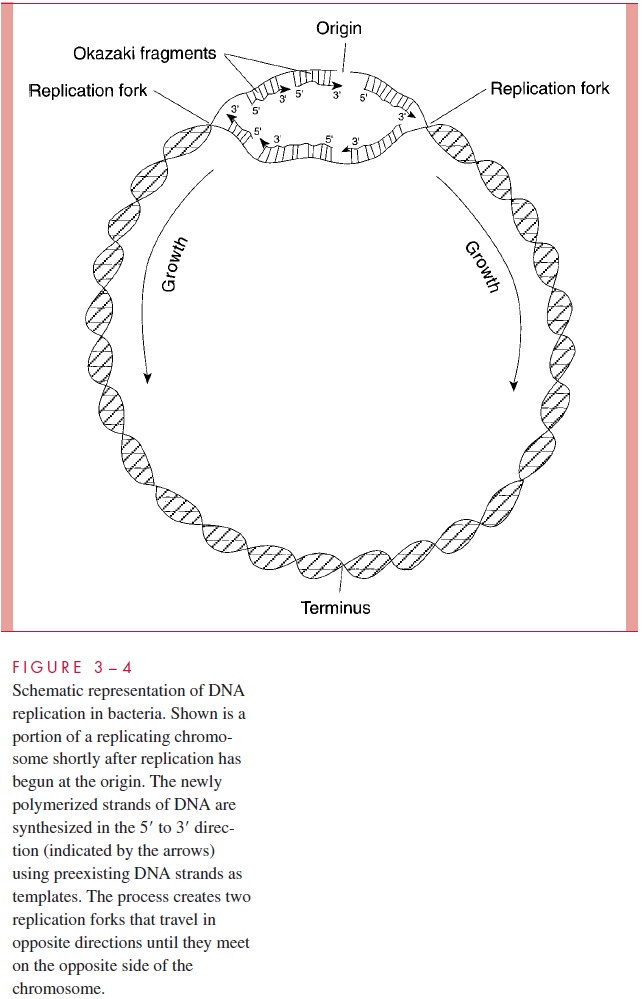
Some chemotherapeutic agents derive their selective toxicity for bacteria from the unique features of prokaryotic DNA replication. The synthetic quinolone compounds in-hibit DNA gyrase, one of the many enzymes participating in DNA replication.
Transcription is the synthesis of RNA. Transcription in bacteria differs from that ineukaryotic cells in several ways. One difference is that all forms of bacterial RNA (mRNA, tRNA, and rRNA) are synthesized by the same enzyme, RNA polymerase. Like the several eukaryotic enzymes, the single bacterial RNA polymerase uses activated building blocks (ATP, GTP, CTP, and UTP) and synthesizes an RNA strand complemen-tary to whichever strand of DNA is serving as template.
A second major difference is that bacterial mRNA need not be transported to the cyto-plasm through a nuclear membrane, and hence no poly(A) cap is needed and no special means of transport exists. In fact, because each mRNA strand is directly accessible to ri-bosomes, binding of the latter to mRNA to form polysomes begins at an early stage in the synthesis of each mRNA molecule (Fig 3 – 5).
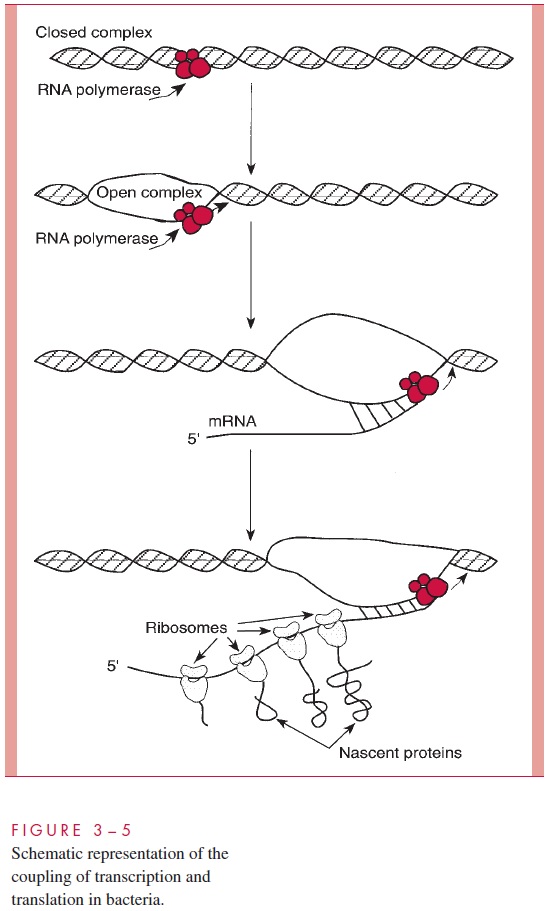
A third remarkable difference is that bacterial mRNA is synthesized, used, and de-graded all in a matter of a few minutes. Although most bacteria have some long-lived species of mRNA, it is characteristic of bacterial cells to “wipe their [transcript] plate clean” every few minutes and make whatever new transcripts are called for by sensing the cell’s environment.
RNA polymerase is a large, complicated molecule with a subunit structure of α2β β’σ . The σ subunit is the one that locates specific DNA sequences, called promoters, which precede all transcriptional units. More than one subunit, each designed to recog-nize a different set of related promoters, can associate with RNA polymerase, which pro-vides a simple means to activate groups of related genes that cooperate in such cellular processes as sporulation, nitrogen acquisition, heat shock stress response, and adaptation to nongrowth conditions.
As in eukaryotic cells, all stable RNA molecules are made from large precursor mole-cules that must be processed by nucleases and then extensively modified to produce the mature product (the tRNAs and rRNAs).
Bacterial RNA polymerase is the target of the rifamycin series of antimicrobics (in-cluding the semisynthetic compound rifampin). They block initiation of transcription. Other substances of biological origin block extension of RNA chains or inhibit transcrip-tion by binding to DNA. They have been of great value in molecular biological studies but are also toxic to human cells and thus are not used in human therapy.
Translation is the name given to protein synthesis. Many antimicrobics derive their se-lective toxicity for bacteria from the unique features of the prokaryotic translation appara-tus. In fact, protein synthesis is the target of a greater variety of antimicrobics than is any other metabolic process . Some agents inhibit the ribosomal large subunit (eg, chloramphenicol and macrolides), some the small subunit (eg, tetracyclines and aminoglycosides), and some aminoglycosides bind to both large and small subunits.
Bacteria activate the 20-amino-acid building blocks of protein in the course of attach-ing them to specific transfer RNA molecules. The aminoacyl-tRNAs are brought to the ri-bosomes by soluble protein factors, and there the amino acids are polymerized into polypeptide chains according to the sequence of codons in the particular mRNA that is being translated. Having donated its amino acid, the tRNA is released from the ribosome to return for another aminoacylation cycle.
This description fits translation in eukaryotic as well as prokaryotic cells, but major differences do exist. The initiation of translation of a new polypeptide chain requires fewer proteins in bacteria. The ribosomes of bacteria are smaller and simpler in structure. Bacterial mRNA is largely polycistronic, that is, each mRNA molecule is the transcript of more than one gene (cistron) and therefore directs the synthesis of more than one polypeptide. No processing or transport of the mRNA is necessary. RNA polymerase makes mRNA at about 55 nucleotides per second (at 37°C), and ribosomes make polypeptide chains at about 18 amino acids per second. Therefore, not only does translation of each mRNA molecule occur simultaneously with transcription, but it oc-curs at the same linear rate (55 nucleotides per second/3 nucleotides per codon 18 amino acids per second). This means that ribosomes are traveling along each mRNA molecule as fast as RNA polymerase makes it. This coupling plays a role in several as-pects of regulation of gene expression unique to bacteria.
These special features of translation in bacteria contribute to the streamlined effi-ciency of the process. The bacterial cytosol is packed with polyribosomes. Each ribosome functions near its maximal rate. Therefore, the faster the growth rate of the cell, the more ribosomes are needed for protein production. It can be estimated that during growth in rich media, more than half the mass of the E. coli cell consists of ribosomes and other parts of the translation machinery.
Other polymerization reactions involve synthesis of peptidoglycan, phospholipid, LPS, and capsular polysaccharide. All of these reactions involve activated building blocks that are polymerized or assembled within or on the exterior surface of the cytoplasmic membrane.
The entire process of synthesizing peptidoglycan (murein), which is completely ab-sent from eukaryotic cells, offers many vulnerable attack points for antibiotics and other chemotherapeutic agents. Some of these are shown in Figure 3 – 6;
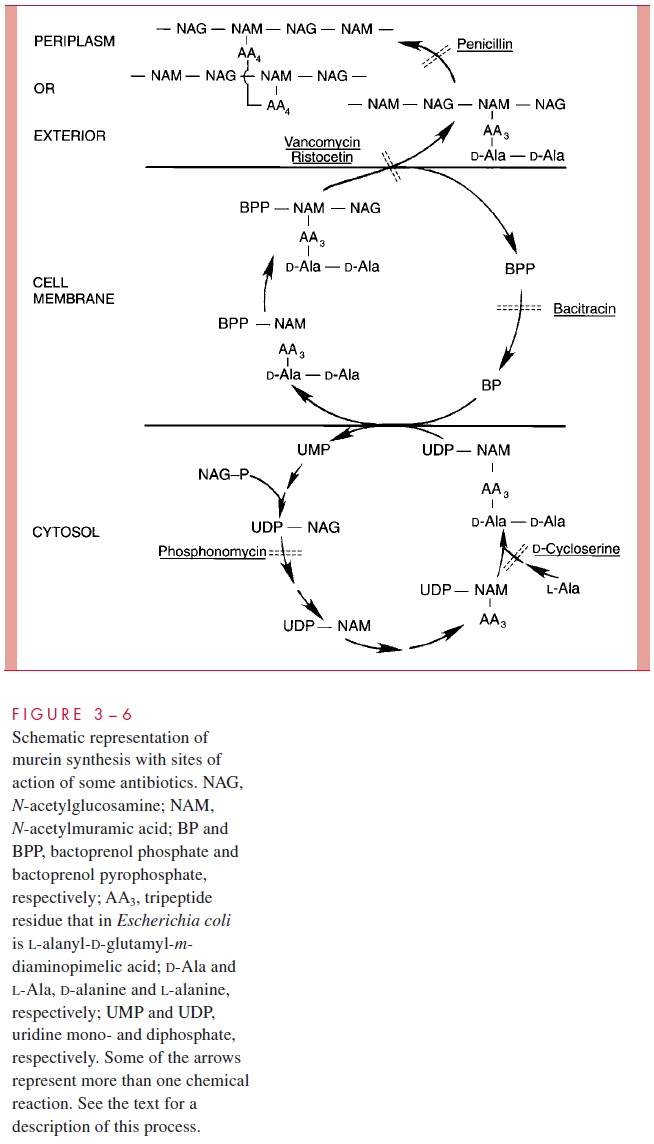
The synthesis of murein occurs in three compartments of the cell (see Fig 3 – 6).
1. In the cytosol a series of reactions leads to the synthesis, on a nucleotide carrier (UDP), of an N-acetylmuramic acid (NAM) residue bearing a pentapeptide (the tetrapeptide found in mature murein plus an additional terminal D-alanine).
2. This precursor is then attached, with the release of UMP, to a special, lipid-like carrier in the cell membrane called bactoprenol (orundecaprenol). Within the cell mem-brane N-acetylglucosamine (NAG) is added to the precursor, along with any amino acids that in this particular species will form the bridge between adjacent tetrapep-tides. Bacitracin and vancomycin interfere with the function of bactoprenol as a car-rier in polymerization and assembly reactions.
3. Outside the cell membrane (in the periplasm of Gram-negative cells and the wall of Gram-positive cells), this disaccharide subunit is attached to the end of a growing gly-can chain, and then cross-links between chains are formed by a transpeptidization us-ing the energy transduced by the release of the terminal D-alanine — the extra aminoacid on the tetrapeptide. Eventually, release from the carrier occurs. These transpepti-dases, called penicillin-binding proteins (PBPs) for their property of combining with this antibiotic, are involved in forging, breaking, and reforging the peptide cross-links between glycan chains. This dynamic process is necessary to permit expansion of the murein sac during cellular growth, to shape the envelope, and to prepare for cell division. It is this process that goes awry in the presence of penicillin and related an-timicrobics, the action of which can be broadly stated as preventing formation of sta-bilizing peptide cross-links.
Related Topics