Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Bacterial Processes
Assembly Reactions and Protein Translocation - Cell Growth of Bacterial
Assembly Reactions and Protein Translocation
Assembly of cell structures occurs both by spontaneous aggregation (self-assembly) and by special, specific mechanisms (guided assembly). Some macromolecules are made at the sites of assembly (such as LPS in the outer membrane), and others must be trans-ported to them (porin is made in the cytosol but ends up in the outer membrane). Self-assembly is illustrated by two cell structures that spontaneously assemble in a test tube from their component macromolecules: flagella and ribosomes. Important parts of enve-lope assembly include special mechanisms for the secretion of proteins, the use of Bayer’s zones of adhesion ( Fig 2 – 9) to form the phospholipid/protein leaflets of the membranes, and the use of carrier molecules (eg, bactoprenol) to transport hydrophilic compounds within the lipid portions of the membrane.
Translocation of Proteins
A problem is posed by the difficulty of moving macromolecules out of the cell interior and into their proper place in the wall, outer membrane, and capsule. Proteins in their natural folded state present a hydrophilic surface that cannot be pushed through phospho-lipid membranes. These proteins may be part of the cell’s assembly process, and are des-tined to reside within the membrane or wall of the cell, or in the case of Gram-negative cells to reside finally in either the periplasm or outer membrane. Moreover, many proteins are translocated through all layers of the cell envelope to the exterior environment. Protein secretion has become the general term to designate all these instances of translo-cation of proteins out of the cytosol (ie, whether the protein is to leave the cell or become part of the envelope), recognizing that all these events share the problem of passing a pro-tein between hydrophilic and hydrophobic phases. An understanding of this complex process is beginning, and it turns out to have great relevance to bacterial virulence. Approximately 20% of the proteins of E. coli are estimated to reside in the envelope. Furthermore, many bacterial virulence factors are located on the surface of the cell, poised to interact with the cells and fluids of the mammalian host. Studies with E. coli and many other Gram-negative as well as Gram-positive bacteria have revealed a surpris-ing number of mechanisms for protein translocation.
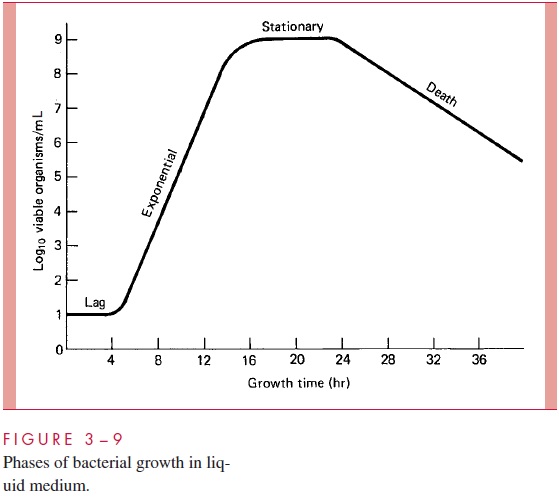
Proteins destined for the wall, membranes, or periplasm are translocated by a generalsecretory pathway (GSP), which consists of cytosolic chaperones and an integral mem-brane translocase consisting of several proteins operating cooperatively. The role of the chaperones is to present the protein to be exported to the translocase, at which a special ATPase “pusher” is thought to physically drive the proteins through the membrane. Pro-teins of the GSP are products of what are called sec (secretory) genes, and the GSP is therefore also called the Sec pathway. Many, but not all, exported proteins are recog-nized by having a special signal sequence at their N-terminus; this peptide is cleaved off during translocation through the membrane by a signal peptidase. The translocation of some proteins occurs cotranslationally (ie, during their synthesis on a ribosome) before the polypeptide has a chance to fold. For some of these, the nascent polypeptide – ribosome complex is docked to the membrane by a signal recognition particle, similar to that in mammalian cells, consisting of a protein (Ffh) and a 4.5S RNA. For others, translocation occurs posttranslationally; the protein is completed and then may be escorted to the translocase by chaperone proteins. Some of these general aspects of protein translocation are shown in Figure 3 – 7.
Export of Proteins
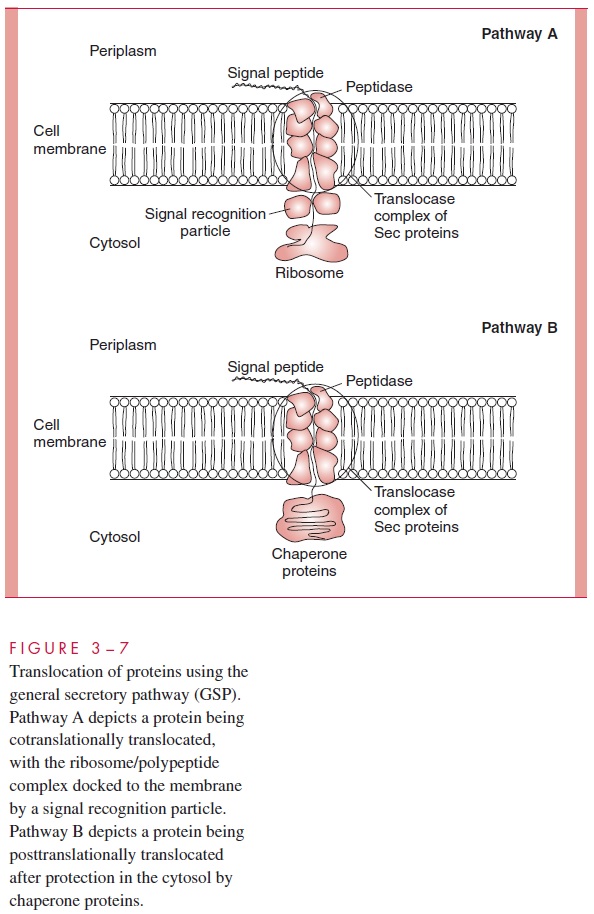
In many cases, proteins are translocated completely through the entire envelope and into the surrounding media or tissue, or even directly into host cells. Secretion of toxins and other proteins contributes greatly to bacterial virulence, and occurs by several pathways, only some of which utilize components of the GSP. In Gram-negative species, secretion must translocate a protein across two membranes. In Gram-positive species, secretion is less complex and usually involves proteins marked by a signal sequence interacting with chaperones and translocases with general similarity to those of the GSP. Five pathways have been discovered in different Gram-negative pathogens that accomplish export of proteins into the environment (Fig 3 – 8). These pathways are important because many of the secreted proteins are toxins or other virulence factors.
Type I secretion systems are Sec-independent (do not use the GSP), and consist of three proteins that form a transmembrane channel through which the secreted protein moves, driven by one of the proteins, an ATP-binding cassette (ABC) transporter; hence, these systems are sometimes called simply ABC transporters. In a single step, the secreted protein, which normally lacks a classical signal sequence, passes from the cytosol to the external environment. The E. coli hemolysin is secreted in this manner.
Type II secretion systems, on the other hand, are Sec-dependent and use the tradi-tional GSP to move a protein into the periplasm, but then in a second step, approximately 14 accessory protein molecules move the secreted protein across the outer membrane. This process is called two-step secretion. Like the type I systems, type II systems in-clude an ATP-binding protein but also a peptidase to cleave a signal sequence from the secreted proteins, all of which have a signal sequence. These systems are common in such Gram-negative bacteria as Klebsiella oxytoca, Vibrio cholerae, Pseudomonas aeruginosa, and E. coli.
Type III systems, which are responsible for the secretion of many virulence factors in Yersinia, Salmonella, Shigella, and Pseudomonas species, involve as many as 20 proteincomponents. One component is a chaperone specific for the given protein to be secreted, and another is an ATP-binding protein thought to energize the system. Type III systems are attracting intense study because they are responsible for contact-dependent secre-tion, in which secretion of virulence proteins is activated by contact with mammalianhost cells, resulting in the direct injection of the secreted protein into the cytoplasm of the mammalian cell. Type III systems are Sec-independent.
Type IV systems are referred to as conjugal transfer systems because they were originally discovered as pathways by which DNA is conjugally transferred between bac-terial cells or between a bacterial and a eukaryotic cell. They are used by the plant pathogen Agrobacterium tumefaciens to transfer oncogenic DNA and protein into plants, and a similar system is used by Bordetella pertussis to export pertussis (whooping cough) toxin. Genes similar to those responsible for this type of secretion in these organisms are found in the pathogenicity island of Helicobacter pylori and inLegionella pneumophila. Currently it is unclear whether the protein secretion by these systems requires the Sec machinery.
Type V secretion systems are two-step, Sec-dependent pathways. No helper protein is needed for translocation through the outer membrane; the transported protein itself accomplishes this feat. Hence, these systems are referred to as autotransporters.
One domain of the protein forms a channel in the outer membrane for the rest of the protein and then is cleaved off. These are the simplest of the Sec-dependent systems. A serine protease from Serratia marcescens, the important IgA proteases of Haemophilus influen-zae and Neisseria gonorrhoeae,and the vacuolating cytotoxin VacA from H. pylori areeach secreted as autotransporters.
Bacterial secretion and export offer potential targets for future design of chemothera-peutic agents.
Related Topics