Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Bacterial Processes
Bacterial Virulence
BACTERIAL VIRULENCE
Special Attributes of Pathogens
Most bacteria have the ability to grow and survive under harsh conditions. Yet of the many thousands of bacterial species, only a small percentage are associated with humans as part of the natural flora or as causative agents of disease. This fact generates the ques-tion central to medical microbiology from the very start: What makes a bacterium patho-genic? The answer is not simple, because it turns out that many properties are necessary for a bacterial cell to gain entrance to a human, evade its defense systems, and establish an infection. They include:
• Adherence to and penetration of host cell surfaces
• Evasion of phagocytic and immunologic attack
• Secretion of toxic proteins to weaken the host and promote spread of the pathogen
• Acquisition of nutrients, including iron, to permit growth within the host
• Survival under adverse conditions both within and outside the host and its macrophages
As we shall see in, the genes unique to pathogens are frequently found clustered in genetic segments within either plasmids or the bacterial chromosome, with interesting implications for the evolution of pathogenic bacteria.
Regulation of Virulence
Most bacterial pathogens must survive and grow in two very different circumstances — in the broad external environment and in or on the human host.
Expressing the very many genes responsible for pathogenesis in the latter situation would be counterproduc-tive, if not outright detrimental to growth and survival in nature, while failing to express them on encountering the host would commonly be fatal. To ensure their growth and survival in both circumstances, bacteria have evolved elaborate and effective mecha-nisms for regulation of virulence genes.
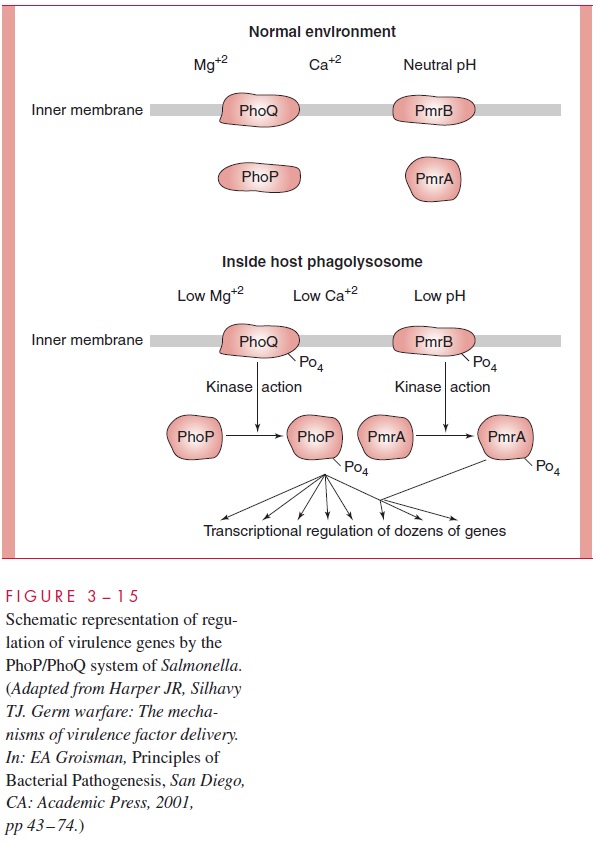
Virulence genes are commonly organized as regulons, and many of these share the attributes of general stress response regulons, described above. For example, many are regulated by classical two-component signal transcription systems in which environmen-tal sensing is achieved by a sensor protein kinase, which relays information about the environment by phosphorylating its partner, a response regulator, which in turn acts to control transcription initiation of its gene set. Many dozens of these systems have been found in various pathogens. Here we examine one example, the PhoP/PhoQ system, which is essential to the virulence of Salmonella (Fig 3 – 15). PhoQ is a sensor protein kinase in the cell membrane, and PhoP is the response regulator to which it relates. PhoQ is sensitive to the concentration of magnesium ion in the periplasm of the Salmo-nella cell. With a normally adequate Mg2+ concentration, PhoQ is locked in an inactivestate; however, when the concentration is very low, as happens when the Salmonella finds itself within the phagolysosome of human macrophages, PhoQ autophosphorylates one of its histidine residues. Phosphorylation of PhoP ensues, and this event activates it as a transcription regulator. The phosphorylated PhoP controls more than 40 genes. Some are induced, including those encoding an acid phosphatase, cation transporters, outer membrane proteins, and enzymes that modify LPS. Some are repressed, including some encoding proteins essential for epithelial cell invasion and others encoding compo-nents of a contact secretion system. The control network is complex, because some of the regulated genes are not directly controlled by PhoP, but by a second two-component response system, called PmrA/PmrB, which is responsive to low pH. The two systems, PhoP/PhoQ and PmrA/PmrB, act in cascade fashion to accomplish a rather intricate re-sponse. The induced proteins are believed to enable the cell to scavenge Mg2+ from its own LPS and to protect itself from the hostile environment of the phagolysosome; the repressed ones were useful in earlier stages of the infective process but now are superflu-ous. The Salmonella cell that finds itself in the phagolysosome of a macrophage has evolved a way to sense its situation and maximize its production of needed factors while dispensing with irrelevant ones.
An interesting principle has emerged from the study of bacterial luminescence and from the field of infectious diseases of plants. Researchers have discovered that several types of bacteria, including Pseudomonas, regulate the expression of genes in a cell density – dependent manner. That is, expression of certain genes occurred only when the population density of the bacteria reached a threshold level, called a quorum. Quorum sensing in some cases is achieved by secretion of a small, diffusible molecule (some are acyl-homoserine lactones) that is sensed by an envelope protein, triggering a regulatory response through a two-component signal transduction system. This autoinduction en-ables the bacteria to avoid “tipping their hand” and mounting an attack on the host before their numbers are sufficient to overwhelm the host’s defenses. Salmonella are known to have a quorum-sensing protein, SdiA, that regulates at least one operon on a virulence plasmid in these cells. No evidence yet indicates the role of this regulation in Salmonella infection. Exploration of the possible role of quorum sensing in general in human disease is ongoing.
Regulation of virulence gene expression is achieved also in a fashion totally unex-pected from the study of metabolic gene regulation, namely by rearrangement of DNA. These instances do not involve mutations in the usual sense of the term, because the DNA alterations are readily reversible. Many well studied examples have generated the concept of a genetic switch, with an “on” and an “off ” position determined by the inversion of a small segment of DNA adjacent to the regulated genes. Examples of this and related mechanisms dependent on DNA recombination.
Related Topics