Chapter: Genetics and Molecular Biology: Attenuation and the trp Operon
RNA Secondary Structure and the Attenuation Mechanism
RNA Secondary Structure and the Attenuation
Mechanism
Point mutations provide excellent support for the
attenuation mechanism as outlined above. Changing the AUG codon of the leader
to an AUA has the expected effect (see Problem 13.2). A base change abolishing
a base pair in the 2-3 hybrid, but one having no effect on the base pairs of
hybrids 1-2 or 3-4 prevents the leader from forming the 2-3 hybrid. Consequently,
the 3-4 hybrid always forms, and termination inevitably results. Mutations that
reduce the stability of the 3-4 hybrid reduce transcription termination, both in vitro and in vivo.
Direct experiments also provide evidence for the
existence of secondary structure in the leader RNA. Light digestion of the RNA
with RNAse T1 cleaves only at those guanosines postulated not to be in the
hybrids (Fig. 13.13). A second experiment also suggests that base pairing in
the leader region is important. As mentioned above, in an in vitro transcription system with purified DNA and RNA
polymerase, virtually all tran-scripts are terminated at the attenuation site.
The substitution of inosine triphosphate (ITP) for GTP however, eliminates the
premature termi-
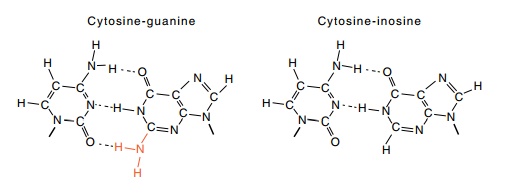
nation. While it is possible that specific contacts
between the inosine and RNA polymerase or between the inosine and the DNA are
respon-sible for the lack of termination, it seems more likely that the failure
of the substituted RNA to form its 3-4 hybrid is the reason. Whereas G-C base
pairs possess three hydrogen bonds, I-C base pairs have only two
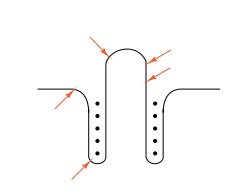
Figure
13.13 The points of RNAseT1 cleavage
of the leader RNA with respect to the 1-2 and 3-4 hybrid structure that likely
forms on na-ked trp leader RNA in vitro.
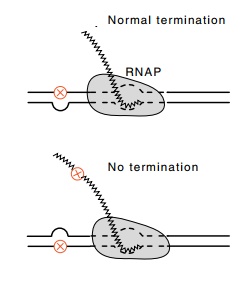
Figure
13.14 The use of hetero-duplex template
DNAs for demonstrating that conforma-tion of the product RNA determines whether
termination occurs. When the mutation is in the strand such that the product
RNA is altered, no termination occurs.
because inosine lacks an amino group that is
present on guanosine. The two hydrogen bonds do not provide sufficient energy
to form the usual hybrids, and thus termination does not occur. Analogs
incorporated into the DNA generate much smaller perturbations on termination,
also suggesting that it is the base-paired structure of the transcribed RNA that
is necessary for termination at the attenuation site. The use of DNA molecules
heteroduplex in the leader region also yields data consistent with the
attenuation mechanism. DNA molecules can be constructed that are wild-type on
one strand and mutant in a single base in the leader region on the other (Fig.
13.14). Only when the mutation is on the strand copied by RNA polymerase and
therefore producing an altered RNA product molecule does the mutant nucleotide
alter attenuation. If the RNA product is wild-type, so is transcription and
termination.
Finally, direct physical evidence for the
attenuation model has also been obtained. In an in vitro transcription system, the usual termination at the
attenuator can be blocked by including a large excess of an oligonucleotide
complementary to region 1. Apparently this oligonu-cleotide anneals to region 1
as it is synthesized and prevents its pairing with region 2. Consequently, the
2-3 hairpin forms, and the 3-4 hairpin, which is required for termination,
cannot form. Therefore, termination does not occur.
Related Topics