Chapter: Genetics and Molecular Biology: Attenuation and the trp Operon
Early Explorations of the Hypersynthesis
Early Explorations of the Hypersynthesis
The experiments described in this section were
performed before the development of modern genetic engineering. It is
interesting to see how Yanofsky and his collaborators cleverly utilized the
technology available at the time to learn the source of the hypersynthesis and
thereby discover the phenomenon of attenuation. The first question in their
careful investigation was whether the hypersynthesis in the internal deletion
strains mentioned above resulted from an alteration in the levels of messenger.
Either more messenger was present in the deletion strains or messenger was
being translated more efficiently. The path of subsequent experiments depended
on which it was.
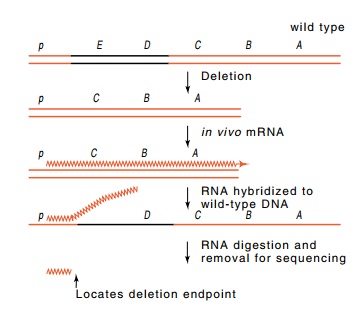
Figure
13.8 RNA synthesizedfrom a deletion,
hybridized to wild-type DNA, and digested with RNAse can reveal the deletion
end point.
Messenger RNA in cells was briefly labeled with tritiated uridine, extracted, and hybridized to complementary trp DNA immobilized on paper. Trp-containing
DNA was available in the form of φ80 and λ phage in which part of the phage genome was
substituted with trp sequences. Use
of such phage containing all or part of the trp
operon substituted for the use of fragments that today would be isolated by
restriction enzyme cleavage of an appropriate plasmid or by PCR amplification
using appropriate primers and crude E.
coli DNA as template. The hybridization revealed that in all strains, the
levels of trpB messenger paralleled
the levels of TrpB enzyme. Therefore the cause of trp enzyme hypersynthesis in some of the internal deletion strains
was elevated messenger levels.
Some of the internal deletion strains
hypersynthesized the TrpB protein and messenger and some did not. Hence it was
important to learn whether the effect could be correlated with the extent of
the deletion. This was particularly important because sequencing of trp messenger had revealed that the trp operon contained an unexpectedly
long leader of 162 bases between the start of transcription and the translation
start of the first trp enzyme.
The deletion end points were determined by
isolating trp messenger synthesized in vivo by the deletion strains. Total
RNA from the deletion strains was isolated and hybridized to wild-type trp DNA (Fig. 13.8). The region of
complementarity between the RNA and DNA was resistant to RNAse. After the
digestion, this region was then melted off the DNA and the exact extent of
trp-specific sequence obtained from each deletion was apparent on RNA
sequencing. RNA sequencing was used since DNA sequencing techniques had not yet
been invented. It was found that only those deletions that removed a site
located 20 nucleotides before the trpE gene
hypersynthesized TrpB protein.
Conceivably, the deletions caused hypersynthesis by
altering the activity of the trp
promoter, although this possibility seemed unlikely because some of the
deletions in the hypersynthesizing strains ended as
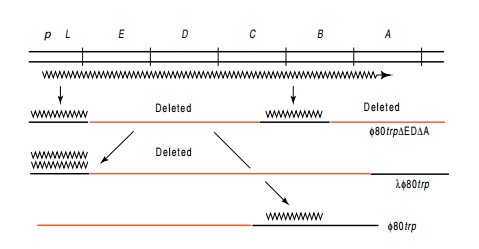
Figure
13.9 Two-step hybridization for
determination of the relative amountsof trpL
and trpB mRNA in cells. Messenger
from the trp operon as indicated on
the top line is hybridized to phage DNA carrying part of the trp operon. RNA eluted from the first
phage was then hybridized to phage containing sequences homologous to either
the leader region or the trpB region
of the operon.
far as
100 nucleotides downstream from the promoter. More likely, the deletions
removed a site located 20 bases ahead of the start of the trpE gene that terminated most transcription before it entered the trp struc-tural genes. This idea was
tested by comparing the amount of trp
messenger specified by a sequence lying upstream of the trpE gene and upstream of the potential termination site to the
amount of messenger specified by sequences downstream from the site (Fig.
13.9). The down-stream messenger contained the trpB region of the operon. A two-step hybridization permitted
quantitation of messenger RNA in cells from each of these regions. This
experiment provided the telling clue. Wild-type repressed cells as well as trpR- cells contained an
eight-to-ten-fold molar excess of messenger from the leader region preceding
the trpE gene over messenger from the
trpB region. In cells deleted of only
the critical region just ahead of the trpE
gene, the amount of trpB messenger
was elevated and was nearly comparable to the amount of trpL RNA. These results prove that, indeed, termination occurs at
the site 20 bases ahead of the trpE
gene and that under normal conditions a majority of the transcription does not
enter the trp structural genes. The
site at which termination occurs is called trp
a.
The obvious and most important question to ask at
this point was whether transcriptional termination, which ordinarily permitted
only one in eight to ten polymerases to transcribe into the trp structural genes, was regulated or
whether the efficiency of termination at trp
a was independent of growth conditions. This was tested by starving for
tryptophan and examining the attenuation level. Suitable starvation was
generated by the addition of indolylacrylic acid that inhibits both tryptophan
synthesis and tryptophanyl-tRNA synthetase. It did decrease termination, and
thus termination at trp a is
regulated, and is called attenuation. Starvation for other amino acids did not
decrease termina-tion at trp a,
thereby proving that termination does play a regulatory role in the trp operon.
In vitro transcription of the trp operon DNA was the logical next stepin these studies. The trp RNA from such experiments could be
quanti-tated by its synthesis from φ80 and λ-trp
phage and appropriate RNA-DNA hybridizations. These showed that virtually all
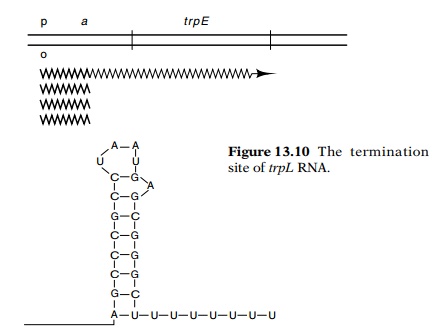
transcripts ended at the point identified in the in vivo experiments as responsible for termination. The sequence of
this region showed that it resembles other sequences at which RNA transcription
terminates (Fig. 13.10). It con-tains a G-C-rich region that can form a hairpin
followed by a string of eight U’s.
The leader region also contains an AUG codon and
can bind ribo-somes just like the beginning of authentic genes. This was shown
by the straightforward but technically difficult experiment of isolating
radio-active leader region RNA from cells, binding ribosomes to it in the
presence of initiation factors and f-met-tRNAfmet,
lightly digesting with RNAse, and sequencing the RNA that was protected from
digestion. Following the AUG codon are 13 more codons before a translation
termination codon. Clearly it was of great interest to determine whether this
leader peptide is synthesized in vivo.
Vigorous attempts to isolate it failed, however. Therefore an indirect
experiment was used to prove that the leader is translated. The leader peptide
region was fused by deletion to the lac
repressor gene, and the corresponding fusion protein was found to be
synthesized at a high rate. Since the leader peptide contains two tryptophans,
termination efficiency appeared to be coupled to translation. This suspicion
was deepened by the findings that trp
regulation is altered in some trp
synthetase mutants.
Related Topics