Chapter: Genetics and Molecular Biology: Regulation of Mating Type in Yeast
Isolation of α2 Protein
Isolation of α2 Protein
The combination of the low synthesis levels of most
regulatory proteins and the fact that few possess any enzymatic activity makes
their detection difficult. The lac
repressor was first detected by virtue of its binding to
iso-propyl-thio-galactoside, IPTG, an inducer of the system that is not cleaved
by β-galactosidase. No small molecule effector of the
mating-type factors is known, and as will be seen below, none is necessary.
Some other method had to be used to identify the α protein.
One approach to the isolation of mating type
encoded factors could be the DNA migration retardation assay using short DNA
fragments containing an appropriate binding site. If the protein binds, the
electro-phoretic migration rate of the fragment is greatly reduced. This assay
is sufficiently selective that it can detect a specific DNA-binding protein
present at only a tiny fraction of total protein. That is, the assay often can
detect a specific DNA-binding protein in crude extracts of the cells. Nowadays
this assay is the method of choice for initial attempts to detect proteins that
bind to DNA.
Since α2 protein
does not come equipped with a convenient enzyme assay and the DNA retardation
assay was not then in wide use, Herskowitz and Johnson used a clever trick for
the isolation of α2. They
fused the entire α2 gene to β-galactosidase. The fusion
protein functions in vivo to repress a-specific genes normally, and theβ-galactosidase alsoretains its activity. Therefore,
the enzyme can be used both to assist the purification by its known behavior in
purification steps, and to assay protein fractions for the hybrid protein
during the purification. Despite susceptibility to proteolytic cleavage in the
cell extracts, the fusion protein could be partially purified by conventional
techniques by track-ing the β-galactosidase
activity through conventional fractionation steps.
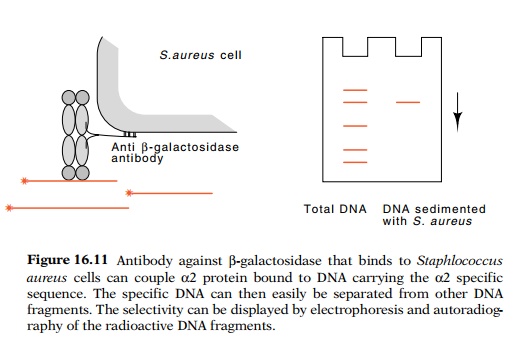
Two assays of DNA binding by α2 fusion protein could be used. The first links a
radioactive DNA fragment containing an α2-binding site to Staphlococcus
aureus cells via antibody andα2-β-galactosidase in aDNA−α2-β-gal-antibody-cell sandwich (Fig.
16.11). The bacteria and their associated complex can be sedimented with a low
speed centrifu-gation and separated from other proteins and from control DNA
frag-ments not containing an α2-binding
site. DNA isolated from the complex was then subjected to electrophoresis to
verify that only the correct DNA fragment was sedimented in the complex.
Another assay for specific binding of α2 protein
was DNAse footprinting.
The binding of α2 to its recognition sequence represses expression of a mating-type genes. Mightα2 be able to repress other genes if itsbinding
sequence were placed in the regulatory region? The answer was yes, but the
results were not simple. We can imagine repression as
resulting from steric exclusion of the binding of
another protein. This was not the case for α2 repressor. Introducing its binding site anywhere upstream of the
cytochrome c gene promoter generated repression. The binding sequence could be
between the enhancer and the TATA box, or even somewhat upstream of the
enhancer element. It did not have to overlap another protein-binding site.
Thus, α2 seems to repress by a different mechanism than by
simple steric exclusion.
Related Topics