Chapter: Genetics and Molecular Biology: Biological Assembly, Ribosomes and Lambda Phage
Experiments with in vitro Ribosome Assembly
Experiments with in vitro Ribosome Assembly
Sophisticated experiments on ribosome structure and
assembly became possible with the ability to isolate individual ribosomal
proteins and to reconstitute ribosomes from them. For example, the
concentration of each protein could be varied in the reconstitution
experiments. This allowed determination of the kinetic order of the reaction,
that is, the number of components that had to interact simultaneously for the
reaction to proceed. It is certainly to be expected that the assembly process
would be sequential, with proteins being added one after an-other; the
alternative case—all 21 of the 30S ribosomal subunit proteins and the rRNA
coming together simultaneously in a reaction involving 22 components—would have
far too low an assembly rate. Indeed, the probability that even a few of the
components come together at once is low. Quite surprisingly, however, the most
rate-limiting step in ribo-some assembly is not the coming together of even two
components; it is the intramolecular rearrangement of a structure already
formed (Fig. 21.5). That is, the rate-limiting reaction is unimolecular, and
its rate relative to the amount of components present cannot be speeded by
increasing the concentration of any components. Such a unimolecular step is
analogous to the isomerization of RNA polymerase to form an open complex at a
promoter.
Figure 21.5 Assembly of 30S subunits. A subset of 30S proteins and 16S rRNAform an RI complex that then must be activated by heat to form RI✵, to which the remainder of the ribosomal proteins bind to form the complete 30S subunit.
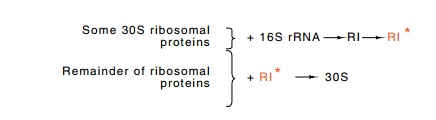
The rate of in
vitro ribosome assembly is strongly temperature-de-pendent. From this
dependence, the activation energy can be calculated to have the unusually high
value of 38 Kcal/mole and is indicative of extensive structural rearrangements.
This high value also suggested that in
vivo ribosome formation also requires high energy. If so, the activa -tion
energy might be increased in some ribosome mutants. This has been found. A
sizable fraction of cold-sensitive mutants in E. coli are defective in
ribosome assembly. At low temperatures, such mutants accumulate precursors of
ribosomes that contain some, but not all, of
Reconstitution of ribosomal subunits from
cold-sensitive mutants permitted pinpointing the target of the mutations. They
were in the ribosomal proteins. A similar approach can also be used to locate
the altered component in other types of ribosome mutations. For example, the
protein that is altered in streptomycin-resistant mutants is a protein
designated S21. The ribosomal proteins are designated by S for proteins of the
smaller, 30S subunit and L for proteins of the larger, 50S subunit. They are
numbered in the order of their locations on a two-dimensional gel in which they
all are separated from one another.
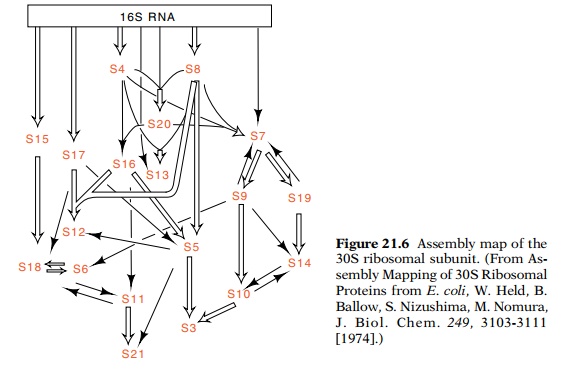
The order that individual ribosomal proteins bind to the maturing ribosome can also be determined by in vitro assembly experiments (Fig. 21.6). The binding of the isolated radioactive
ribosomal proteins to rRNA can be measured by sedimentation of the RNA after
incubation. Only a subset of the 30S proteins are capable of binding to the
naked 16S rRNA. The complexes thus formed can then be used to determine which
proteins can bind next. Building in this fashion, a complete assembly map of
the ribosomal subunits has been constructed. In general it shows that any of
several proteins can bind to the structure at any stage in its assembly. Also,
a few of the proteins that interact strongly during assembly are encoded by
genes in the same ribosomal protein operon (Fig. 21.7).
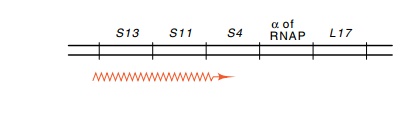
Figure 21.7 Gene structure of the ribosomal protein operon containing S13,S11, and S4. This operon also contains the gene for the sigma subunit of RNA polymerase and the ribosomal protein L17.
Related Topics