Chapter: Genetics and Molecular Biology: Generating Genetic Diversity: Antibodies
Engineering Antibody Synthesis in Bacteria
Engineering Antibody Synthesis in Bacteria
There are limitations to polyclonal and monoclonal
antibodies that can be found. Antibodies desired for special purposes, for
example investi-gation of enzyme catalysis, could require antibodies of such
selectivity that few are likely to be found by conventional methods. While
animals may synthesize hundreds of antibodies against a large antigen, the
number is limited by the size of the B cell population. Even somatic mutation can
be imagined not to generate all desired structures in antigen-binding sites of
antibodies. How could vast numbers of variant antibodies be generated for the
study of antibody-antigen interactions?
One method for increasing the number of antibody
specificities available for study would be to express a large library of light
and heavy chains in bacteria. If these large libraries could be combined, then
the same multiplicative increase in diversity as occurs in nature could also be
available. Furthermore, expressing the proteins in bacteria simplifies
Figure
20.13 Amplification of an mRNA sequence
coding for an immuno-globulin gene. Reverse transcriptase is used at the first
step to make DNA. Thereafter the thermoresistant Taq polymerase is used in normal PCR reac-tions. The sequences
encoding the restriction enzyme cleavage sites do not hybridize to the template
on the first cycle. Thereafter they do.
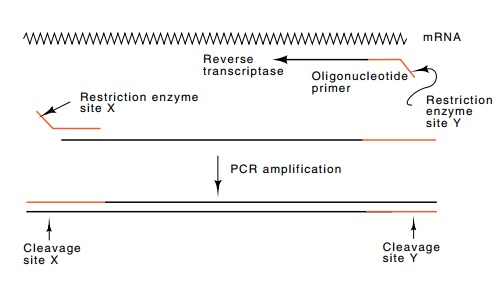
How might one begin to express a wide collection of
light or heavy chains in bacteria? We cannot simply take a collection of V
region genes, for most in the genome are not connected to the D or J regions
and no diversification has occurred at the splicing step. We need to clone
after rearrangement has taken place, and it would be best to clone only genes
that have rearranged. If, however, we clone from DNA, the intervening sequences
still interrupt the genes. The best approach is to clone from the mRNA using
reverse transcriptase to make DNA copies. Then the polymerase chain reaction
can be used to amplify the DNA. Due to the high fraction of conserved amino
acids in the variable and light chain regions, it is possible to design
oligonucleotides that will specifically hybridize to regions of the genes. The
oligonucleotides can also be made with restriction enzyme cleavage sites so
that later the resulting DNA can be easily inserted into a cloning vector (Fig.
20.13).
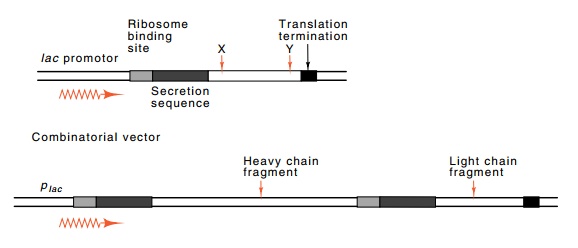
Cleverness
in designing the cloning vector also is helpful. The incom-ing DNA must be
expressed; therefore the plasmid must contain an active promoter. It should be
possible to regulate this promoter in case the gene products are lethal to the
cells. The lac promoter is a good one
to use. Downstream from the promoter must be a good ribosome-bind-ing site.
Beyond this it is helpful to have a sequence that will direct the resulting
protein to be secreted from the cells. Then, at last, comes the cloning site
which is flanked by the two restriction enzyme cleavage sites. The same sites
were engineered into the DNA that was amplified by PCR. Finally, a translation
termination sequence is needed. Addi-tional factors are that after generating
libraries of light and heavy chains, combinations of the two must be formed.
Resulting vectors must carry one light and one heavy chain with both being
expressed under control of the lac
promoter (Fig. 20.14).
Screening
of the recombined library for vectors carrying a light and heavy chain that
bind a particular antigen must be easy. Techniques have been developed for
screening large numbers of lambda plaques on plates, and thus the best vector
for initial screening would be lambda phage rather than a plasmid. Later, once
a desired recombinant has been found, the light and heavy chains should be
transferred to a plasmid vector. Finally, rather than attempting to synthesize
the entire antibody molecule, just the Fab’ fragment should be sufficient for
many studies, and the probes for PCR amplification can be designed
accordingly.
The
technology described above has worked well and libraries have been made
containing up to 106 different light chains and 106
different heavy chain fragments. These have been combined to yield libraries
with truly gargantuan numbers of different antibody specificities.
Related Topics