Chapter: Genetics and Molecular Biology: Generating Genetic Diversity: Antibodies
D Regions in H Chains
The D Regions in H Chains
The heavy chains of mice and humans have also been
examined to determine whether they are spliced together from segments as are
the light chains. Indeed, the heavy chains are. About 300 variable H chain
segments were found as well as four J regions in mice and ten in humans. There
also exists a stretch of C regions, one gene for each of the constant region
classes. These segments, however, do not encode all the amino acids found in
the heavy chains. Thus it seemed likely that
an additional segment of DNA is spliced in to
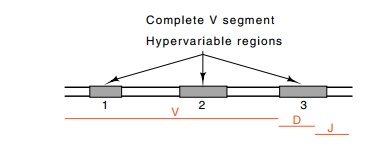
generate the complete immunoglobulin heavy chain. These segments are called D,
for diver-sity, because they lie in the third hypervariable region of the heavy
chain.
Almost the same strategy as was used to find the J
regions worked in finding the missing segments of the heavy chains. Tonegawa
had ob-served that the J regions in some myelomas were rearranged even though
they were not connected to V segments. Most likely the rear-rangements involved
the D segments. This hypothesis proved to be true. By cloning a rearranged J
region and using the D region it contained, the D regions were located on the
embryonic DNA ahead of the J regions.
One of the interesting features of the D regions is
that they possess flanking sequences of seven and nine bases separated by 12 or
23 base pairs (Fig. 20.9) as do the sequences and spacers found alongside the V
and J regions of the light chain. These flanking sequences are recognized by
the DNA rearranging system. Cutting and joining is between a pair of sequences,
one possessing a 12 base spacer, and one containing a 23 base spacer.
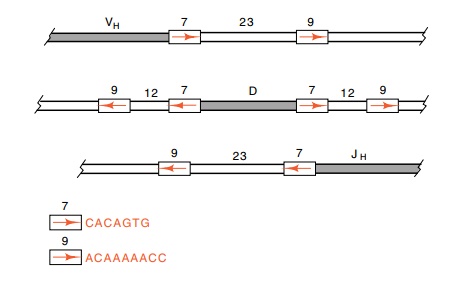
Figure
20.9 The V, D, and J regions and the
positions, sizes, and sequences ofthe flanking recognition sequences used in the
processing to produce an intact immunoglobulin gene.
Not only does introduction of J and D regions
generate diversity, but imperfections in the rearranging process itself also
generate diversity. That is, the cutting and joining are not perfect. Often deletions
or insertions of several bases are made as the D segment is spliced into
position (Fig. 20.10).
The variable region of the H chain that is
generated by the fusions and rearrangements of different DNA regions is joined
to the C region.
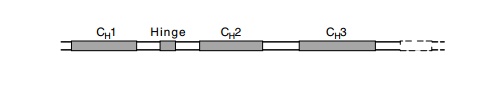
The C region of the heavy chain itself contains
three intervening se-quences. One lies between the coding region for each
stretch of 100
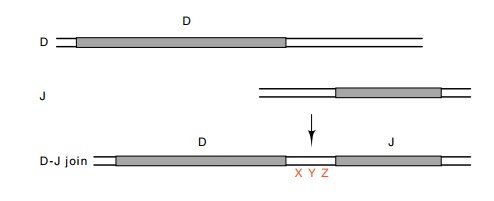
Figure
20.10 An insertion of three bases
generated upon splicing a D and J segment.
amino acids. These groups of 100 amino acids form
the domains of the protein that interact with the portions of the immune system
that dispose of foreign molecules bound to antibody molecules. The fact that
intervening sequences separate well-defined structural domains of a protein led
to the prediction mentioned earlier that most intervening sequences in other
proteins would also be found to separate the nucleo-tides coding for functional
and structural domains of the protein. Such a spacing increases the chances
that sequences coding for intact do-mains could be shuffled between proteins in
the course of evolution.
Related Topics