Chapter: Genetics and Molecular Biology: Generating Genetic Diversity: Antibodies
Assaying for Sequence Requirements of Gene Rearrangements
Assaying for Sequence Requirements of Gene Rearrangements
It is not easy to assay for the sequence requirements of the joining signals used in the assembly of the immunoglobulins. Many copies of these sequences exist in normal cells and it is impossible to change all of them and then measure the new rates of recombination. An additional difficulty in the study of the recombination process is the low rate of recombination. Such low rates are necessary so that two recombination events do not often simultaneously occur in one cell.
To assay for recombination we need a method that will permit alteration of the relevant sequences and permit us to look at just the genes that are recombined. One way is to design a vector that can be transformed into cell lines established from different cell types of the immune system. The DNA could be left a while in the cells, then removed and transformed into E. coli, where we want to be able to detect easily just those rare DNA molecules that have been acted upon by the
Figure 20.15 Recombinational joining between signal sequences generates acoding joint and a signal joint.
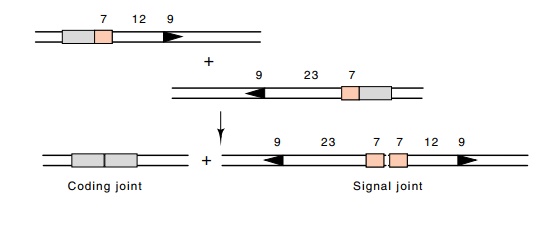
Sequence analysis has shown that the recombinant junction signals always recombine a signal with a 12 base spacing with a signal contain-ing a 23 base spacing (Fig. 20.15). Two products result, a coding joint and a signal joint. Part of the diversity of the immune system is the inaccuracy that is generated at the coding joint. Nucleotides are deleted and inserted at this point. Very few such inaccuracies occur at the signal junction. If one of the junction signals were inverted, then, just like integration and excision of phage lambda, the DNA lying between the coding regions would not be deleted, but would be inverted.
The inversion of a DNA segment can be utilized in the design of a vector to assay for recombinational joining. By designing the vector to generate inversions rather than deletions, the products of both the coding joint and the signal joint can be preserved for further study. This can be done by arranging that a promoter on the vector be prevented from driving expression of a drug resistance gene by the presence of a transcription terminator between the two (Fig. 20.16). Flanking the terminator are a pair of recognition sequences oriented so that recom-bination inverts the DNA, thereby nullifying the terminator and permit-ting transcription of the drug resistance gene.
Figure 20.16 Recombination between the signal sequences inverts the regionbetween the two, thereby inactivating the terminator by turning it backwards and upside-down. This activates expression of tetracycline resistance.
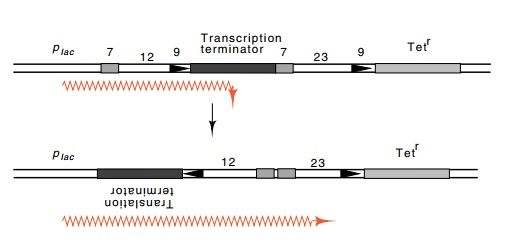
Samples of the plasmid DNA with the promoter, terminator, and recognition sequences, can be transformed into cells, allowed to rest there for a long period, extracted, and then transformed into bacteria. The total number of transformed bacteria can be assayed by the number of colonies receiving the plasmid, and the fraction which are also tetracycline resistant provides a measure of the recombination which occurred while the DNA was within the animal cells. The results of such experiments show that bases near the coding splice site are most important, as are the spacing distances.
Related Topics