Chapter: Biotechnology Applying the Genetic Revolution: Inherited Defects
Dominant Mutations May Be Positive or Negative
DOMINANT MUTATIONS MAY BE POSITIVE OR NEGATIVE
Occasionally a mutation
causes a change of function or even a gain of function in the resulting gene
product. In this case, a single mutant copy of the gene may cause significant
phenotypic effects—that is, the mutation is dominant. Gain-of-function
mutations are relatively rare in inherited disease but are frequent in the
somatic mutations that cause cancer.
Gain-of-function mutations
are much more specific than those causing loss of function. For example,
achondroplasia, or short-limbed dwarfism, is due to mutation of a single copy
of the FGFR3 gene that encodes fibroblast growth factor (FGF) receptor #3. FGF
receptors are signal-transducing proteins, that is, they receive signals at the
cell membrane and transmit the information to the nucleus. Normally they are
activated only when they receive a signal from outside the cell (i.e., bind
FGF). Although most mutations inactivate the receptor, a few extremely rare
mutations yield receptors that are active despite the absence of an external
signal. Only mutations that cause the replacement of the amino acid glycine at
position 380 with arginine (Gly380Arg)
cause achondroplasia. Other mutations in this gene cause other symptoms.
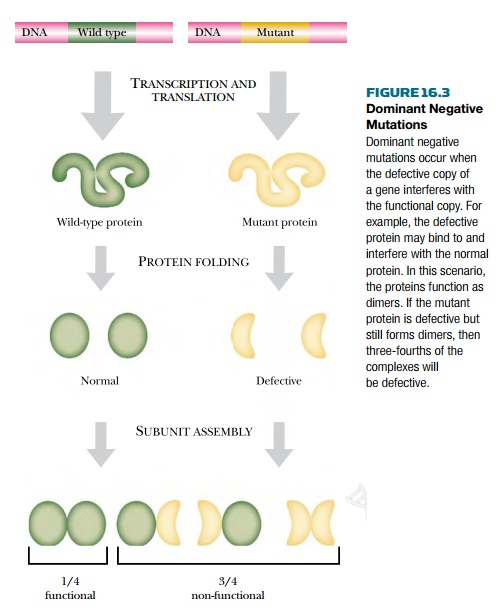
Dominant negative mutations
are those where the mutant protein loses its own function but, in addition, the
defective protein interferes with the function of another protein. Thus a
dominant negative mutation usually results from the presence of an altered,
defective protein. Mutations in the same gene that merely abolish protein
synthesis are usually recessive.
The simplest scenario for a
dominant negative mutation is where the affected protein forms oligomers. If a
mutation blocks protein synthesis in one copy, there will simply be a 50%
reduction in the amount of protein. However, if the mutant allele produces an
altered protein, this may bind to normal copies of the same protein and give
inactive complexes.
The final result may be that
almost no active protein is available (Fig. 16.3). Many transcription factors
bind to DNA as dimers and are susceptible to dominant negative effects.
Proteins that form ion channels are also liable to dominant negative effects.
Cardiac arrhythmia (Romano-Ward syndrome) is due to a dominant mutation in the
KVLQT1 gene. This encodes a protein that assembles with others to form a
transmembrane potassium channel. Certain defective KVLQT1 proteins can still
bind as part of the assembly but block the operation of the channel. About 20%
remaining activity is seen in this syndrome.
Related Topics