Chapter: Biochemistry: Nucleic Acid Metabolism
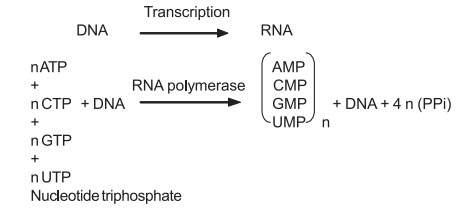
Biosynthesis of RNA (Transcription)
Biosynthesis of RNA (Transcription)
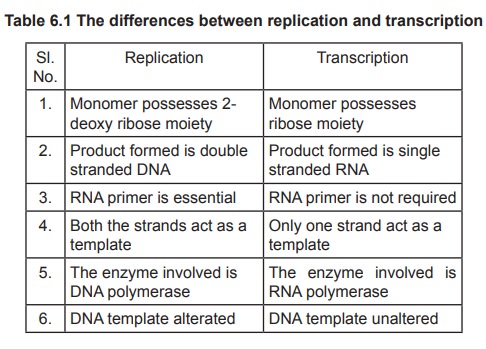
The biosynthesis of RNA is very much similar to
that of DNA, except that in RNA it is differed by having different RNA types
(mRNA, tRNA, rRNA), and the nitrogen base uracil instead of thymine. Like DNA,
polymerization of 4 nucleoside triphosphates (viz ATP, CTP, GTP and UTP) in the
presence of Mg2+ (or) Mn2+ ion is catalysed by the enzyme RNA polymerase and
one strand of DNA serves as template.
Mechanism
Transcription involves 3 stages
i. Initiation
ii. Elongation
iii.
Termination
Three phases of transcription
i. Initiation
In E.
coli, all genes are transcribed by a single large RNA polymerase. This
complex enzyme, called the holoenzyme is needed to initiate transcription since
the factor is essential for recognition of the promoter. It is common for
prokaryotes to have several s factors that recognize different types of
promoter (in E.coli, the most common
s factor is σ70).
The holoenzyme binds to a promoter region about
40-60 bp in size and then initiates transcription a short distance downstream
(i.e. 3' to the promoter). With in the promoter lie two 6 bp sequences that are
particularly important for promoter function and which are therefore highly
conserved between species. Using the convention of calling the first nucleotide
of a transcribed sequence as +1, there 2 promoter elements lie at position -10
and -35, that is about 10 and 35 bp respectively, downstream of where
transcription will begin.
ii. Elongation
After transcription initiation, the s factor is
released from the transcriptional complex to leave the core enzyme (α2ββ'ω)
which continues elongation of the RNA transcript. Thus, the core enzyme
contains the catalytic site for polymerisation, probably within the subunit.
The first nucleotide in the RNA transcript is usually PPPG (or) PPPA. The RNA
polymerase then synthesises the RNA in the 5' -> 3' direction, using the 4
ribonucleotide 5' triphosphates (ATP, CTP, GTP, UTP), as precursors.
The 3'-OH group at the end of the growing RNA
chain attaches to a phosphate group of the incoming ribonucleotide 5'
triphosphate to form a 3',5' phosphodiester bond (figure 6.8). The complex of
RNA polymerase, DNA template and new RNA transcript is called a ternary complex
(i.e three components) and the region of unwound DNA that is undergoing
transcription is called transcription bubble (figure 6.9). The RNA transcript
forms a transient RNA-DNA hybrid helix with its template strand but then peels
away from the DNA as transcription proceeds. The DNA is unwound ahead of the
transcription bubble and after the transcription complex has passed, the DNA
rewinds.
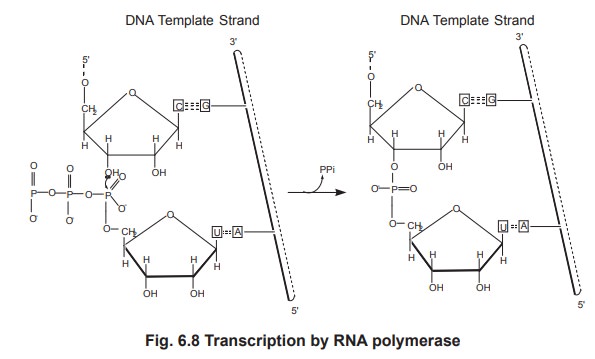
In each step the incoming ribonucleotide
selected is that which can base pair with the next base of the DNA template
strand. In the diagram, the incoming nucleotide is UTP to base pair with the A
residue of the template DNA. A 3',5' phosphodiester bond is formed, extending
the RNA chain by one nucleotide, and pyrophosphate is released. Overall the RNA
molecule is grown in a 5' to 3' direction.
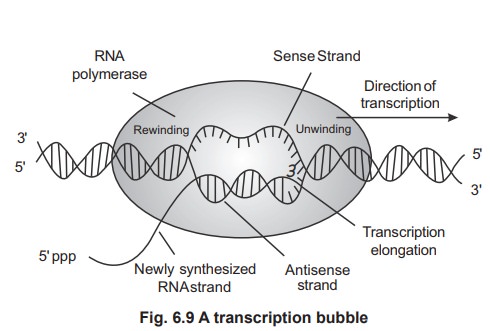
The DNA double helix is unwound and RNA
polymerase then synthesizes an RNA copy of the DNA template strand. The nascent
RNA transiently forms an RNA-DNA hybrid helix but then peels away from the DNA
which is subsequently rewound into a helix once more.
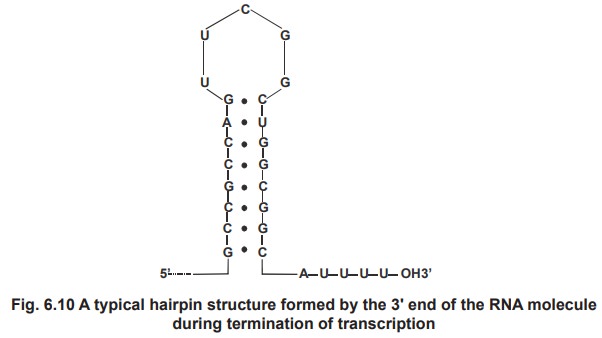
3. Termination
Transcription continues until a termination
sequence is reached. The most common termination signal is a G == C rich region
is a palindrome, followed by an A = T rich sequence. The RNA made from the DNA
palindrome is self complementary and so base pairs internally to form a hairpin
structure rich in GC base pairs followed by four or more U residues (fig.6.10).
However not all termination sites base this hairpin structure. Those that lack
such a structure require an additional protein, called rho protein (r) to help
recognize the termination site and stop transcription.
Post Transcriptional Modification
Post-transcriptional modications are not needed
for prokaryotic mRNAs. The formation of functionally active RNA molecule
continues after transcription is completed in eukoryotes. The product of
transcription in eukarryotes are called as primary transcripts and they undergo
modification by a process called post transcriptional modification.
1. Processing of mRNA molecules
mRNA undergoes several modifications before it
is being translated. They are
1.
Addition
of blocks of poly A, containing 200 (or) more AMP residues to the 3' end of
messenger RNAs. This addition of poly A tail takes place in the nucleus.
2.
A
methylated Guanine nucleotide called as “5' cap” is added to the 5' end of mRNA
molecule.
3.
Methylation
occurs in the internal adenylate nucleotides at their N-6 position. It’s
function is not known.Thus mRNA is processed to get the active mRNA molecule.

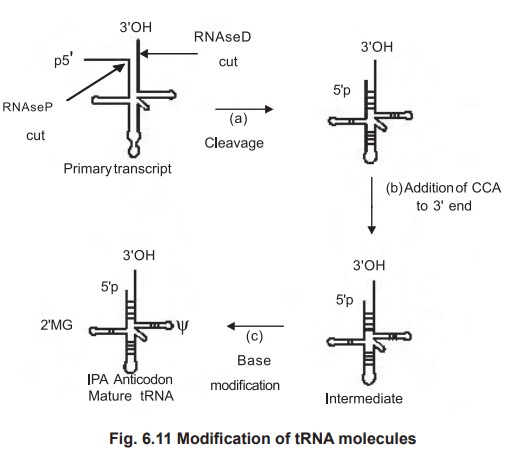
2. mRNA Processing of tRNA Molecules
Most cells have 40 to 50 distinct tRNAs.
Transfer RNA’s are derived from longer RNA precursors of enzymatic removal of
extranucleotide units from the 5' and 3' ends.
1. Formation of the 3'-OH terminus
This process involves the action of an
endonuclease that recognizes a hairpin loop at the 3' end called RNAse D, which
stops two bases at short of CCA terminus, though it later removes these two
bases after the 5' end is processed. This enzymatic digestion leaves the
molecule called pre-tRNA.
2. Formation of the 5'-P terminus
The 5'-P terminus is formed by an enzyme called
RNAse P, which removes excess RNA from the 5' end of a precursor molecule by an
endonucleolytic cleavage that generates the correct 5' end.
3. Production of modified bases
The final modification is to produce the altered
bases in the tRNA.
In tRNA, two uridines are converted to pseudo
uridine (Ψ), one guanosine to methyl guanosine (MG), one adenine to
isopentyladenine (IPA) etc.
Role of tRNA in protein synthesis
Transfer RNA is the smallest polymeric RNA.
These molecules seem to be generated by the nuclear processing of a precursor
molecule.
The tRNA molecules serve a number of functions,
the most important of which is to activate amino acids for protein synthesis.
The function of tRNA is to bind the specific aminoacids, one might think that
there are 20 types of tRNAs (i.e as many as the constituent amino acids of
proteins). Since the code is degenerate (i.e. there is more than one codon for
a aminoacid). There are also more than one tRNA for a specific aminoacid.
Therefore there are generally several tRNAs specific for the same aminoacid
(Sometimes upto 4 or 5); they are called isoacceptor tRNA’s. These various
tRNA’s, capable of binding the same amino acid, differ in their nucleotide
sequence, they can either have the same anticodon and therefore recognize the
same codon (or) have different anticodons and thus permit the incorporation of
the aminoacid in response to multiple codons specifying the same aminoacid.
As mentioned, each tRNA is specific for an
aminoacid i.e. it can bind (or accept) only that particular aminoacid. Thus,
tRNAAla denotes a tRNA specific for alanine.
Codon is made up of 3 bases, and is present in
mRNA. The tRNA contains anticodon which is complementary (opposite) to codons
in mRNA.
Related Topics