Chapter: Biochemical Pharmacology : Some principles of cancer pharmacotherapy
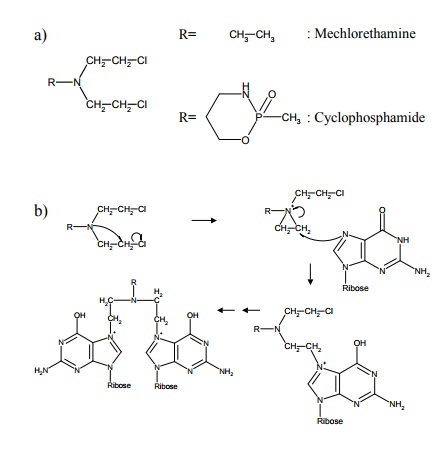
Alkylating agents
Alkylating agents
Synthetic
drugs that are not cell cycle-specific are mostly alkylating agents. They have
diverse reactive groups. Sev-eral drugs share the `N-mustard' structure shown
in Figure 13.7a. The reaction mechanism (formation of aziridine in-termediates
that react as electrophiles; Figure 13.7b) is ac-tually the same as previously
discussed for the irreversible α-blocker phenoxybenzamine; here, however, we
don't have a moiety that targets the drug to any particular protein. It
should go without saying that most molecules will actu-ally not react with DNA
but instead with some other nucle-ophile hopping about in the cell, in
particular glutathione or other sulfhydryls. However, those that do react with
DNA are the ones that matter, since the harm done by them has the potential to
be permanent. Also note that we have not one but two chloroethyl groups – this
creates the possibili-ty of introducing cross-links into the DNA7.
Cross-links be-tween the two strands of the DNA bases are more likely to give
rise to permanent mutations than modifications affect-ing one strand only,
since they cannot be removed by exci sion repair (although there are
`error-prone repair' mecha-nisms that may remove them).
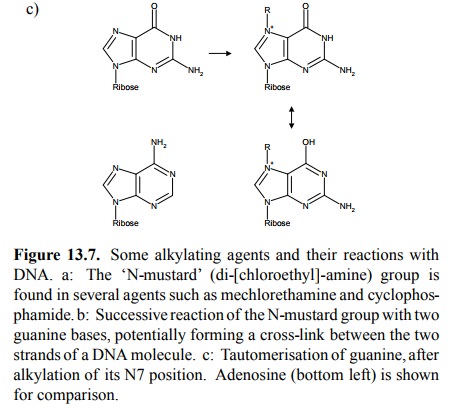
The most common target of
alkylation – quite indepen-dently of the alkylating agent used – is the N7 in
guanine (Figure 13.7b,c). Why is that? The six-membered ring that is part of
the guanine is locked up in base pairing within the double helix, i.e.
inaccessible; this lets out the other ni-trogens. The same applies to the other
nucleotides. Why would N7 in guanine be more commonly affected than N7 in
adenine? The guanine ring is not as completely aromat-ic as the adenine ring
is. The π electrons of the ring nitro-gens are therefore not as completely
delocalized, i.e. the nitrogens will be stronger nucleophiles. However, the
pref erence is not absolute, and alkylations of adenine and the pyrimidine
bases do occur as well.
An interesting consequence of
guanine N7-alkylation is the increased propensity of the guanine ring to adopt
the tau-tomeric form (Figure 13.7c). In this form, the arrangement of hydrogen
bond donors and acceptors is reversed and now resembles that of adenine, thus
enabling guanine to base pair with thymine instead of cytosine. This is
believed to contribute to the mutagenic effect of guanine alkylation.
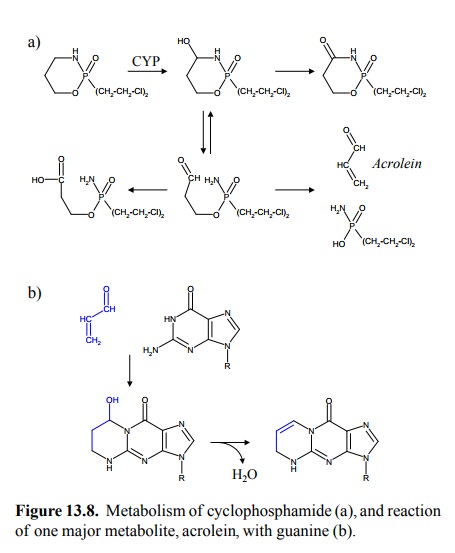
One very
commonly used agent containing the di-chloroethyl-amine moiety is
cyclophosphamide. This drug may actually be metabolized quite extensively and
give rise to several toxic metabolites, the exact contribution of which to the
overall therapeutic effect is not very well established (Figure 13.8a). Metabolism
is initiated by a cytochrome P450 enzyme in the liver (and possibly elsewhere)
and continued by several other enzymatic and non-enzymatic steps. One of the
final decay products is acrolein 8, which may form several adducts
with guanine, some of which are shown in Figure 13.8b. While the metabolites of
cy-clophosphamide such as acrolein and chloroacetaldehyde seem to be
quantitatively more important in the ultimate reactions with DNA than the
parent drug itself, the relative significance of individual metabolites is
somewhat hard to determine from the literature.
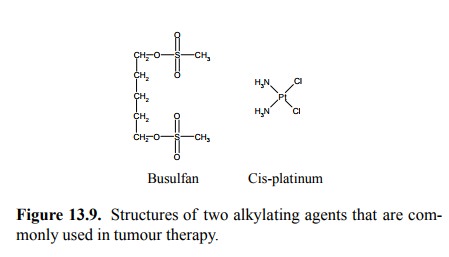
Alkylating agents with other
reactive groups do exist – e.g., busulfan and cis-platinum (Figure 13.9) – but
we will light-ly gloss over their respective intricacies, in particular the
chemical mechanism of cisplatinum and only note that the preferentially formed
lesion observed with the latter drug again involves guanines; two that are
located adjacently within the same strand become cross-linked to each other.
What are
the biological consequences of the covalent mod-ifications caused by alkylating
agents? One that we have already noted is the introduction of mutations
opposite a modified base. Another one is the inhibition of DNA syn-thesis;
e.g., some of the acrolein adducts of guanine (Fig-ure 13.8b) inhibit the
incorporation of any base opposite to them, thus interfering with DNA synthesis
and repair. Of note, many of these chemical adducts are removed by DNA repair
enzymes only inefficiently or not at all; the enzymes are apparently not `accustomed'
to these peculiar types of modifications.
Related Topics