Chapter: Genetics and Molecular Biology: Generating Genetic Diversity: Antibodies
Structure of Antibodies
The Structure of Antibodies
Determining the amino acid sequences and
three-dimensional structures of antibodies has greatly helped our understanding
of how they are encoded by a reasonable number of genes and how they function.
Although eight major classes of human antibody are known, this chapter deals
primarily with the IgG group which contains IgG3, IgG1, IgG2b, and IgG2a. This is
the most prevalent antibody in serum. The other antibodies are IgM, the first
detectable antibody synthesized in response to an antigen, IgD, IgE, and IgA.
These have slightly different structures and apparently are specialized for
slightly different biological roles. IgG contains four polypeptide chains, two
identical light, or L, chains of 22,500 molecular weight, and two identical
heavy, or H, chains of about 50,000 molecular weight, giving a structure H2L2. Two
subclasses of light chains are known in mouse and humans, kappa and lambda,
whereas the eight major immunoglobulin classes are determined by the eight
types of heavy chain, M, D, G3, G1, G2b, G2a, E, and A.
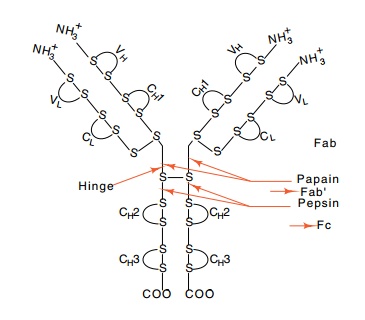
The chains of IgG are linked by inter- and
intrachain disulfide bonds into a Y-shaped structure (Fig. 20.3). Papain
digestion of one mole of IgG yields two moles of Fab fragments and one mole of
Fc fragment. The “ab” indicates that these fragments possess the same binding
specificity as the intact IgG antibody, and the “c” indicates that these fragments
are readily crystallizable. Analysis of fragments of IgG has
Figure
20.3 The structure ofIgG. The
antigen-binding site is at the amino terminal end. V and C indicate variable
and constant regions, respectively. H and L indicate heavy and light chains,
respectively, and the numbering refers to do-mains.
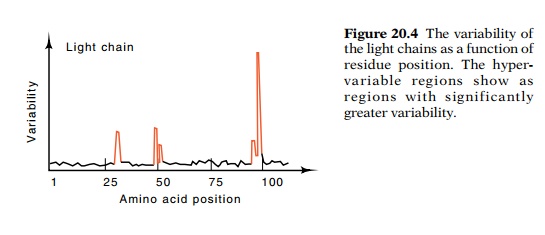
The amino acid sequences of both the L and the H
chains of IgG isolated from myelomas revealed an astonishing fact. About the
first 100 amino acid residues of each of these chains are highly variable from
antibody to antibody, whereas the remainder of the chains is constant (Fig.
20.4). Even more remarkable, however, is the finding that three or four regions
within the variable portion of the protein are hypervariable. That is, most of
the variability of the proteins lies in these regions. In view of their
variability, such regions are likely to constitute the anti-gen-binding site of
the antibody molecule. This suspicion was confirmed by affinity labeling of
antibodies with special small-molecule antigens that chemically link to nearby
amino acid residues. They were found to link to amino acids of the hypervariable
portions of the L and H chains, just as expected.
Proteins from myelomas also assisted the X-ray
crystallography of antibodies. Without the homogeneity provided by monoclonal
antibod-ies, the crystal structures of antibodies could never have been deter
Figure
20.4 The variability ofthe light
chains as a function of residue position. The hyper-variable regions show as
regions with significantly greater variability.
Figure
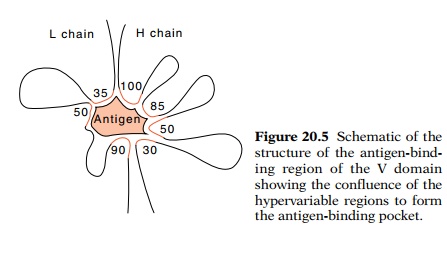
20.5 Schematic of thestructure of the
antigen-bind-ing region of the V domain showing the confluence of the
hypervariable regions to form the antigen-binding pocket.
The structure of Fab fragments confirms most of the postulates that the amino
acid sequence raised. The hypervariable portions of the L and H chains together
form the antigen-binding site at the end of each arm of the antibody molecule
in a domain formed by the variable regions (Fig. 20.5). In addition, the constant
region of the L chain and the first constant region, CH1, of the H
chains form another domain (shown earlier in Fig. 20.3). The constant regions CH2
and CH3 of the two heavy chains form two additional domains of
similar structure.
The fact
that the variable portions of the L and H chains together form the
antigen-binding site greatly reduces the number of genes required to specify
antibodies. If any one of 1,000 L chains could combine with any one of 1,000
different H chains, then a total of 2,000 different genes could code for
antibodies, with a total of 1 × 106
different binding specificities. The next sections describe how B lymphocytes
combine small numbers of segments of the L and H genes to generate large
numbers of different L and H genes, which then generate still larger numbers of
antibodies because an L chain can function with most H chains. Techniques
similar to those used for study of the antibody genes from B cells have also
been applied to the identification and analysis of clones containing the genes
encoding the T cell antigen receptors. These proteins also contain two
polypeptides with variable and constant regions, and DNA rearrangements during
the ontogeny of the organism also generate diversity.
Related Topics