Chapter: Biochemistry: Transcription of the Genetic Code: The Biosynthesis of RNA
Post transcriptional RNA Modification
Post transcriptional RNA
Modification
The
three principal kinds of RNA-tRNA, rRNA, and mRNA-are all modified
enzymatically after transcription to give rise to the functional form of the
RNA in question. The type of processing in prokaryotes can differ greatly from
that in eukaryotes, especially in the case of mRNA. The initial size of the RNA
transcripts is greater than the final size because of leader sequences at the
5' end and trailer sequences at the 3' end. The leader and trailer sequences
must be removed, and other forms of trimming
are also possible. Terminal sequences
can be added after transcription, and base
modification is frequently observed, especially in tRNA.
Transfer RNA and Ribosomal RNA
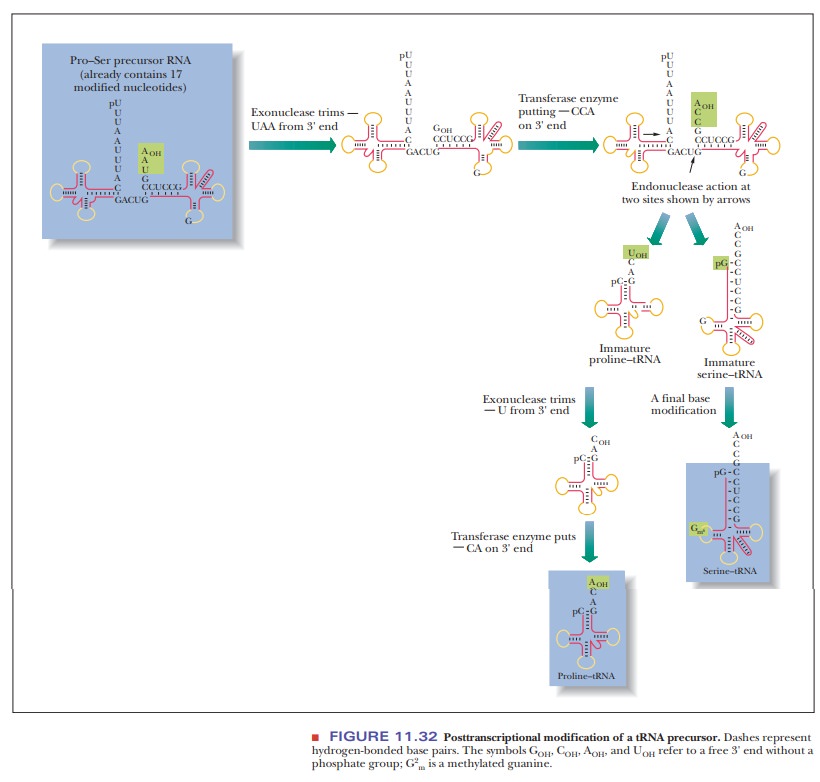
The
precursor of several tRNA molecules is frequently transcribed in one long
polynucleotide sequence. All three types of modification-trimming, addition of
terminal sequences, and base modification-take place in the transformation of
the initial transcript to the mature tRNAs (Figure 11.32). (The enzyme
responsible for generating the 5' ends of all E. coli tRNAs, RNase P,
consists of both RNA and protein.) The RNA moiety is responsible for the
catalytic activity. This was one of the first examples of catalytic RNA. Some
base modifications take place before trimming, and some occur after.
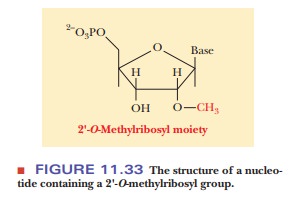
Methylation and substitution of sulfur for oxygen are two of the more usual
types of base modification. One type of methylated nucleotide found only in
eukaryotes contains a 2'-O-methylribosyl
group (Figure 11.33).
The
trimming and addition of terminal nucleotides produce tRNAs with the proper
size and base sequence. Every tRNA contains a CCA sequence at the 3' end. The
presence of this portion of the molecule is of great importance in protein
syn-thesis because the 3' end is the acceptor for amino acids to be added to a
growing protein chain. Trimming of large precursors of eukaryotic tRNAs takes
place in the nucleus, but most methylating enzymes occur in the cytosol.
The
processing of rRNAs is primarily a matter of methylation and of trimming to the
proper size. In prokaryotes, there are three rRNAs in an intact ribosome, which
has a sedimentation coefficient of 70S. In the smaller subunit, which has a
sedimentation coeffi-cient of 30S, one RNA molecule has a sedimentation
coefficient of 16S. The 50S subunit contains two kinds of RNA, with
sedimentation coefficients of 5S and 23S. The ribosomes of eukaryotes have a
sedimentation coefficient of 80S, with 40S and 60S subunits. The 40S subunit
contains an 18S RNA, and the 60S subunit contains a 5S RNA, a 5.8S RNA, and a
28S RNA. Base modifications in both pro-karyotic and eukaryotic rRNA are
accomplished primarily by methylation.
Messenger RNA
Extensive
processing takes place in eukaryotic mRNA. Modifications include capping of the 5' end, polyadenylating (adding a poly-A
sequence to) the 3'end, and splicing
of coding sequences. Such processing is not a feature of the synthesis of
prokaryotic mRNA.
Why is mRNA modified after initial transcription?
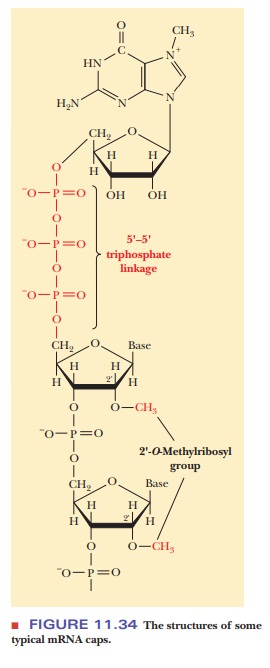
The cap at the 5' end of eukaryotic mRNA is a guanylate residue that is methylated at the N-7 position. This modified guanylate residue is attached to the neighboring residue by a 5'-5' triphosphate linkage (Figure 11.34). The 2'-hydroxyl group of the ribosyl portion of the neighboring residue is frequently methylated, and sometimes that of the next nearest neighbor is as well.
The polyadenylate tail (abbreviatedpoly-Aorpoly[r(A)n]) at the 3' end of a message(typically 100 to
200 nucleotides long) is added before the mRNA leaves the nucleus. It is
thought that the presence of the tail protects the mRNA from nucleases and
phosphatases, which would degrade it. According to this point of view, the
adenylate residues would be cleaved off before the portion of the molecule that
contains the actual message is attacked. The presence of the 5' cap also
protects the mRNA from exonuclease degradation.
The
presence of the poly-A tail has been very fortuitous for researchers. By
designing an affinity chromatography column
with a poly-T tail (or
poly[d(T)] tail), the isolation of mRNA from a cell lysate can be quickly
accomplished. This enables the study of transcription by looking at which genes
are being transcribed at a particular time under various cell conditions.
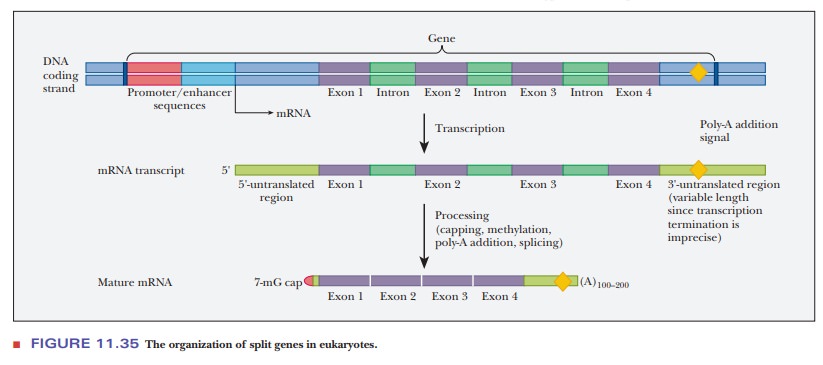
The
genes of prokaryotes are continuous; every base pair in a continuous
prokaryotic gene is reflected in the base sequence of mRNA. The genes of
eukaryotes are not necessarily continuous; eukaryotic genes frequently contain
intervening sequences that do not appear in the final base sequence of the mRNA
for that gene product. The DNA sequences that are expressed (the ones actually
retained in the final mRNA product) are called exons.The inter-vening sequences, which are not expressed, are
called introns. Such genes are often
referred to as split genes. The
expression of a eukaryotic gene involves not just its transcription but also
the processing of the primary transcript into its final form. Figure 11.35
shows how a split gene might be processed. When the gene is transcribed, the
mRNA transcript contains regions at the 5' and 3' ends that are not translated
and several introns shown in green. The introns are removed, linking the exons
together. The 3' end is modified by adding a poly-A tail and a 7-mG cap to
yield the mature mRNA.
Some
genes have very few introns, while others have many. There is one intron in the
gene for the muscle protein actin; there are two for both the α- and β-chains of hemoglobin, three for lysozyme, and
so on, up to as manyas 50 introns in a single gene. The pro α-2 collagen gene in chickens
is about 40,000 base pairs long, but the actual coding regions amount to only
5000 base pairs spread out over 51 exons. With so much splicing needed, the
splicing mechanisms must be very accurate. Splicing is a little easier because
the genes have the exons in the correct order, even if they are separated by
introns. Also, the primary transcript is usually spliced in the same positions
in all tissues of the organism.
A major exception to this is the splicing that occurs with immunoglobulins, in which antibody diversity is maintained by having multiple ways of splicing mRNA. In the last few years, more eukaryotic proteins that are the products of alternative splicing have been discovered. The need for this was also demon-strated by the preliminary data from the Human Genome Project. Differential splicing would be necessary to explain the fact that the known number of pro-teins exceeds the number of human genes found.
How are introns spliced out to make mature RNA?
The Splicing Reaction: Lariats and Snurps
The
removal of intervening sequences takes place in the nucleus, where RNA forms ribonucleoprotein particles (RNPs)
through association with a set of nuclear proteins. These proteins interact
with RNA as it is formed, keeping it in a form that can be accessed by other
proteins and enzymes. The substrate for splicing is the capped, polyadenylated
pre-mRNA. Splicing requires cleavage at the 5' and 3' end of introns and the
joining of the two ends. This process must be done with great precision to
avoid shifting the sequence of the mRNA product. Specific sequences make up the
splice sites for the process, with GU
at the 5' end and AG at the 3' end of the introns in higher eukaryotes. A branch site within the intron also has a
conserved sequence. This site is found 18 to 40 bases upstream from the 3'
splice site. The branch site sequence in higher eukaryotes is PyNPyPuAPy, where
Py represents any pyrimidine and Pu any purine. N can be any nucleotide. The A
is invariant.
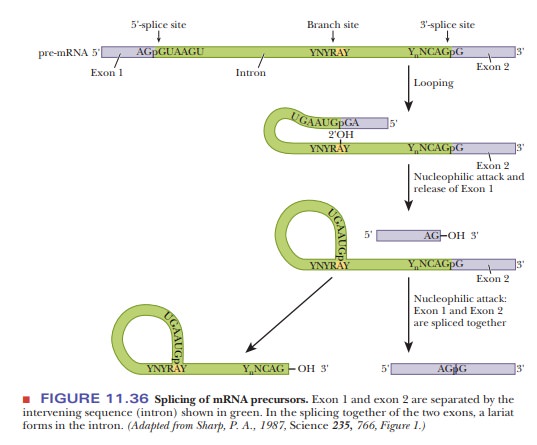
Figure
11.36 shows how splicing occurs. The G that is always present on the 5' end of
the intron loops back in close contact with the invariant A from the branch
point. The 2' hydroxyl of the A performs a nucleophilic attack on the
phosphodiester backbone at the 5' splice site, forming a lariat structure and releasing exon 1. The AG at the 3' end of the
exon then does the same to the G at the 3' splice site, fusing the two exons.
These lariat structures can be seen with an electron microscope, although the
structure is inherently unstable and soon is linearized.
The splicing also depends on small nuclear ribonucleoproteins, or snRNPs (pronounced “snurps”), to mediate the process. This snRNP is another basic type of RNA, separate from mRNA, tRNA, and rRNA. The snRNPs, as their name implies, contain both RNA and proteins. The RNA portion is between 100 and 200 nucleotides in higher eukaryotes, and there are 10 or more proteins.
With
more than 100,000 copies of some snRNPs in eukaryotic cells, snRNPs are one of
the most abundant gene products. They are enriched in uridine residues and are
therefore often given names like U1 and U2. snRNPs also have an internal
consensus sequence of AUUUUUG. The snRNPs bind to the RNAs being spliced via
complementary regions between the snRNP and the branch and splice sites. The
actual splicing involves a 50S to 60S particle called the spliceosome, which is a large multisubunit particle similar in size
to a ribo-some. Several different snRNPs are involved, and there is an ordered
addition of them to the complex. In addition to their role in splicing, certain
snRNPs have been found to stimulate transcription elongation. It is now widely
recog-nized that some RNAs can catalyze their own self-splicing, as will be
discussed. The present process involving ribonucleoproteins may well have
evolved from the self-splicing of RNAs. An important similarity between the two
processes is that both proceed via a lariat mechanism by which the splice sites
are brought together. The following Biochemical Connections box describes an
autoimmune disease that develops when the body makes antibodies to one of these
snRNPs.
Alternative RNA Splicing
Gene
expression can also be controlled at the level of RNA splicing. Many proteins
are always spliced in the same way, but many others can be spliced in different
ways to give different isoforms of
the protein to be produced. In humans, 5% of the proteins produced have
isoforms based on alternative splicing. These differences may be seen by having
two forms of the mRNA in the same cell, or there might be only one form in one
tissue, but a different form in another tissue. Regulatory proteins can affect
the recognition of splice sites and direct the alternative splicing.
It has been found that a protein called Tau accumulates in the brain of people with Alzheimer’s disease. This protein has six isoforms generated by differential splicing, with the forms appearing during specific developmental stages. The human troponin T gene produces a muscle protein that has many isoforms because of differential splicing. Figure 11.37 shows the complexity of this gene.
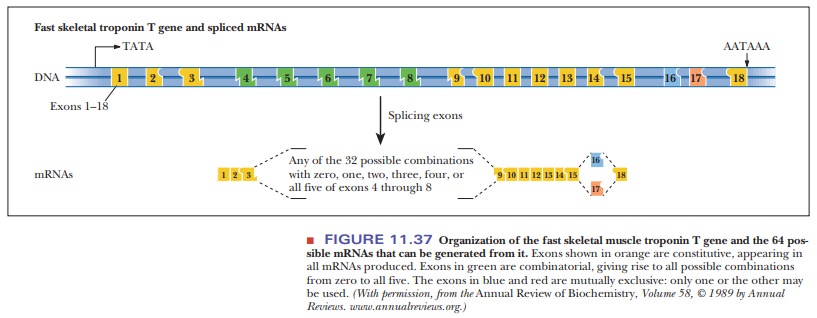
Eighteen exons can be linked together to make the mature mRNA. Some of
them are always present, such as exons 1–3 and 9–15, which are always linked
together in their respective orders. However, exons 4–8 can be added in any
combination group of 32 possible combinations. On the right side, either exon
16 or 17 is used, but not both. This leads to a total of 64 possible troponin
molecules, which highlights the tremendous diversity in protein structure and
function that can come from splicing mRNA.
Summary
After being transcribed from DNA, many RNA
molecules are modified, often extensively, before they arrive at their final
form.
Several modifications are common with tRNA and
rRNA, such as trim-ming, addition of terminal sequences, and base modification.
Messenger RNA is modified by putting a cap on
the 5' end and a poly-A tail on the 3' end.
Messenger RNA is also modified by the removal
of intervening sequences, or introns.
The reaction that removes
introns involves the formation of a lariat as an intermediate. Splicing also
depends on a separate type of RNA called a small nuclear ribonucleoprotein, or
snRNP (“snurp”).
Alternative splicing of mRNA helps account for
the fact that there are more proteins produced in eukaryotes than there are
separate genes.
Related Topics