Chapter: Genetics and Molecular Biology: Lambda Phage Genes and Regulatory Circuitry
Entropy, a Basis for Lambda Repressor Inactivation
Entropy, a Basis for Lambda Repressor Inactivation
The binding of dimeric repressor to operator can be
described by a dissociation constant that is related to the change in the free
energy via the standard thermodynamic relationship:
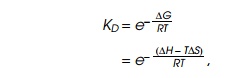
where KD is the dissociation constant of the dimer from
operator, DG is the change in free energy, R is the gas constant, T
is the temperature, DS is the
change in entropy, andDH is the
change in enthalpy.
It is
natural to assume that if monomeric repressor were binding to operator, then
roughly half as many contacts would be formed between
repressor
and DNA and roughly half as many water molecules and ions would be displaced
from DNA and the protein as the repressor bound. Therefore we might write for
the dissociation constant of the monomer where KM is the dissociation constant for monomer,
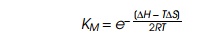
or KM= KD1/2.
This is not correct, however. To make this clear, let DS be
written as the sum of the entropy changes involved with the contacts and
displacement of water, DSinteraction
dimer plus the change in entropy involved with
immobilization and orientation of the dimeric repressor,
DSintrinsic
dimer:
DSdimer=DSinteraction dimer+DSintrinsic dimer.
The same
type of equation can be written for the monomer:
DSmonomer=DSinteraction monomer+DSintrinsic monomer.
Roughly
the monomer makes half as many interactions as the dimer and displaces half as
many water molecules and ions as it binds. Therefore
DSinteraction monomer=DSinteraction dimer/2.
The same is not true of the intrinsic entropies.
The entropy change associated with bringing repressor monomer to rest on
operator by correctly positioning and orienting it is nearly the same as the
change associated with the dimeric repressor. Thus
DSmonomer≠DSdimer/2,
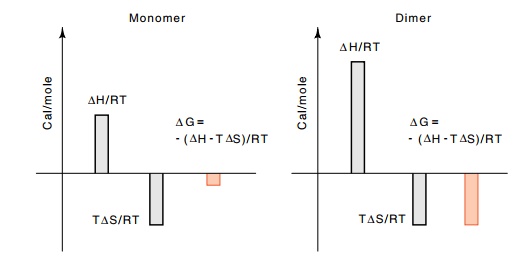
Figure
14.18 Why a dimer can bind much more
tightly than a momomer.Almost all the additional DH provided by the second monomer’s binding can go into increasing DG, whereas for binding of a monomer, almost no DG is left over to contribute to binding.
and
detailed calculations based on statistical mechanics or experiments show that
the inequality can be severe. This is another example of the chelate effect
discussed earlier. In other words, a sizable fraction of the total entropy
change involved with repressor binding to DNA is associ-ated with its correct
positioning. Roughly the same entropy is required to orient a monomer or dimer,
but in the case of lambda repressor, the presence of the second subunit of the
dimer adds twice as much to the binding energy. Most of this additional energy
can go into holding the dimer on the DNA, and hence KD is much less than K2M (Fig. 14.18).
Related Topics