Chapter: Genetics and Molecular Biology: Lambda Phage Genes and Regulatory Circuitry
Initiating DNA Synthesis with the O and P Proteins
Initiating DNA Synthesis with the O and P
Proteins
Transcription
originating from pR leads
to the accumulation of the O and P proteins. These, plus transcription into or
near the ori site, initiate a cascade
of assembly and disassembly whose function is to denature the origin region.
Denatured origin is the target of DNA primase and its availability permits
replication to begin.
When the
O protein has reached appropriate levels, it binds to a series of four repeats
of a palindromic DNA sequence located at ori
(Fig. 14.7). After the O protein has bound, the P protein with bound host
protein DnaB binds to O protein. The host chaperonin proteins DnaJ and DnaK,
that were mentioned earlier for their roles in renaturing denatured proteins,
release P protein and the helicase activity of DnaB plus torsion on the DNA
generated by transcription nearby helps separate the strands at the origin.
With the additional assistance of the host proteins, DnaE, DnaG, DnaZ, RNA
polymerase, and DNA gyrase, DNA synthesis initiates and proceeds outward in
both directions from ori. Ordinarily
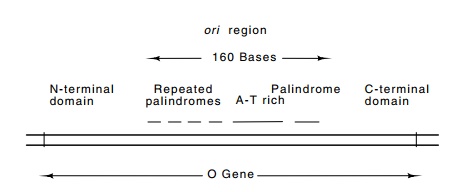
Figure
14.7 The structure and location of theoriregion within the lambdaOgene.
the
requirement for transcriptional activation of ori is met by transcrip-tion originating from pR. Other lambda mutants may be isolated that create
other promoters that activated ori.
Their transcription does not have to cross ori.
One mutant promoter that activates lambda phage DNA replication was 95 base
pairs away, and its transcription is directed away from ori.
Curiously,
ori is located within the lambda O gene itself! The coding regions of the
O gene on either side of ori specify well-defined domains. The
amino acids encoded by the portion of O
containing ori–the four repeats, an
A-T-rich region, and a palindrome–are not predicted to possess much secondary
structure and are very sensitive to proteases. Such a picture is consistent with
the experimental findings that the N-terminal portion of the O protein contains
the phage-specific DNA determinants and that the carboxy-terminus of O binds
the lambda P protein. The O protein itself is highly unstable in vivo.
Mutations
in the ori site itself support the
structure described above. Ori mutants
are recognized because they are cis-dominant
mutationsaffecting DNA replication and they generate very tiny plaques. The
sequence of several such mutations has shown that they lie in the ori section of O. Some are small deletions within the ori region, but they all preserve the reading frame; that is, they
are multiples of three bases. Therefore, the amino acids encoded by the ori portion of the O gene are unimportant to the functioning of O protein.
Related Topics