Chapter: Genetics and Molecular Biology: Lambda Phage Genes and Regulatory Circuitry
Genetic Structure of Lambda
The Genetic Structure of Lambda
A turbid or opaque lawn of cells is
formed when 105 or more cells are spread on the agar surface in a
petri plate and allowed to grow until limited by nutrient availability. If a
few of the original cells are infected with a virus, severalcycleof lysis of
the infected cells and infection of adjacent cells during growth of the lawn
leaves a clear hole in the bacterial lawn. These holes are called plaques. The
first mutations isolated and mapped in phage lambda were those that changed the
morphology of its plaques. Ordinarily, lambda plaques are turbid or even
contain a minicolony of cells in the plaque center. Both result from growth of
cells that have become immune to lambda infection. Conse-quently, lambda
mutants that do not permit cells to become immune produce clear plaques. These
may easily be identified amid many turbid plaques. Kaiser isolated and mapped
such clear plaque mutants of lambda. These fell into three complementation
groups, which Kaiser called CI, CII, and CIII, with the C standing for clear.
One
particular mutation in the CI gene is
especially useful. It is known as CI857, and the mutant CI product is temperature-sensitive. At
tem-peratures below 37° the
phage forms normal turbid plaques, but at temperatures above 37° the mutant forms clear plaques.
Furthermore, lysogens of lambda CI857 can be induced to switch from
lysogenic to lytic mode by shifting their temperature above 37°. They then excise from the
chromosome and grow vegetatively.
Many
additional lambda mutations were isolated and mapped by Campbell. These could
be in essential genes, as he used conditional mutations. Such mutants are
isolated by plating mutagenized phage on a nonsense-suppressing strain. Plaques
deriving from phage containing nonsense mutations may be identified by their
inability to grow after being spotted onto a nonsuppressing strain.
Phage
with nonsense mutations in various genes may then be studied by first preparing
phage stocks of the nonsense mutants on suppressing hosts. The phage can then
be used in a variety of studies. For example, pairs of mutants can be crossed
against one another and the frequency
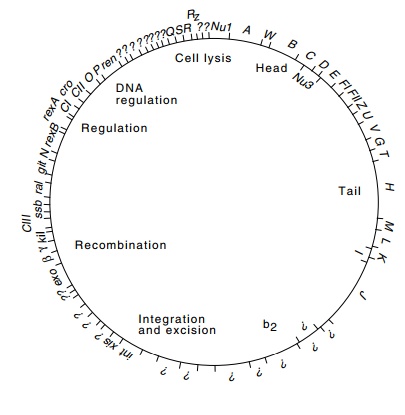
Figure
14.2 The complete physi-cal map of
lambda determined from its DNA sequence. The sizes of genes are indicated along
with the functions of various classes of proteins. The genetic map is simi-lar
except that the circle is opened between genes Rz and Nu1.
of generation of wild-type phage by recombination
between the two mutations can be quantitated by plating on nonsuppressing
strains. In this way a genetic map can be constructed. The nonsense mutants
also facilitate study of phage gene function. Nonsuppressing cells infected
with a nonsense mutant phage stock progress only partway through an infective
cycle. The step of phage development and maturation that is blocked by the
mutation can be determined with radioactive isotopes to quantitate protein,
RNA, and DNA synthesis, or electron microscopy to determine which phage
macromolecules or structures are synthesized.
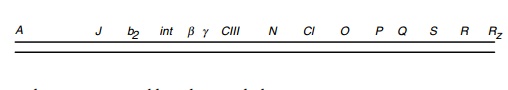
Campbell named the genes he found and mapped A
through R, left to right on the genetic map. The genes identified and mapped
after his work are identified by the remaining letters of the alphabet, by
three-
The
sequence of the entire lambda DNA molecule has been deter-mined. A few of the
known genes could be identified in the sequence by the amino acid sequences of
their products or by mutations that changed the DNA sequence. Many others could
be identified with a high degree of confidence by the correspondence between
genetic and physi-cal map location (Fig. 14.2). The identification of such open
reading frames was greatly assisted by examination of codon usage. An open reading
frame that is not translated into protein usually contains all sense codons at
about the same frequency, whereas the reading frames that are translated into
protein tend to use a subset of the codons. That is, many proteins use certain
codons with a substantially higher fre-quency than other codons. Unexpectedly,
applying these criteria to the DNA sequence of phage lambda revealed more than
ten possible genes not previously identified genetically or biochemically. It
remains to be shown how many of these actually play a role in phage growth and
development.
The DNA
sequencing revealed a second unexpected property of the lambda genes. Many are
partially overlapped. The function of this overlap could be to conserve coding
material, although it may also play
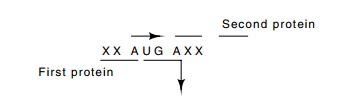
a role in
translation. Since ribosomes drift phaselessly forward and backward after
encountering a nonsense codon and before dissociating from the mRNA, this
overlapping may help adjust the translation efficiency of the downstream gene
product.
Genes of
related function are clustered in the lambda genome. The genes A through F are required for head formation, and Z through J for tail
formation. The genes in the b2
region are not essential for phage growth under normal laboratory conditions
and may be deleted without material effect. Since such deletion phage still
possess the normal protein coat but lack about 10% of their DNA, they are less
dense than the wild-type phage; that is, they are buoyant density mutants. The
genes int and xis code for proteins that are involved with integration
andexcision and will be discussed in a later chapter. The genes exo, β, and γ are involved with recombination.
The proteins from genesCIII,N,CI,cro, and CII are expressed early in the growth cycle and regulate theexpression
of phage genes expressed early. The genes O
and P code for proteins required for
initiation of lambda DNA replication. The Q
gene product regulates expression of late genes. These genes include S, R,
and Rz, which are required
for lysis of the cell as well as the genes that encode proteins comprising the
head and tail of the phage particle.
Related Topics