Chapter: Genetics and Molecular Biology: Xenopus 5S RNA Synthesis
Biology of 5S RNA Synthesis in Xenopus
Biology of 5S RNA Synthesis in Xenopus
Oocytes develop in the ovaries of female frogs in a
development process that takes from three months to several years. Eventually
the oocytes mature as eggs ready to be fertilized. These huge cells contain
large amounts of nutrients and protein synthesis machinery so that later, the
fertilized egg can undergo many cell divisions without significant intake of
nutrients and without additional ribosome synthesis.
Curiously, during oogenesis components of the
ribosomes are not synthesized in parallel. In the ovaries the 5S RNA components
of ribosomes are synthesized first, for about two months. As they are
synthesized, they are stored away in two types of RNA-protein particles.
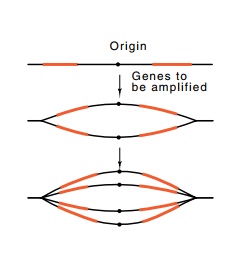
Figure
15.1 Two rounds of initia-tion from an
origin can yield four copies of genes immediately sur-rounding that origin.
These are known as 7S and 42S particles from their
rates of sedimenta-tion in centrifugation. Even though 5S RNA synthesis
proceeds for a long time, the enormous number of RNA molecules needed in an egg
necessitates that multiple genes encode the RNA. The Xenopus genome contains about 100,000 copies of the 5S gene. A
special RNA polymerase transcribes these genes as well as genes coding for
tRNAs. This is RNA polymerase III. Although it is different from the two other
cellular RNA polymerases, it shares a number of subunits in common with them.
After synthesis of the 5S RNA, synthesis of the
18S, 28S, and 5.8S ribosomal RNAs and the ribosomal proteins begins. We might
expect the frog genome also to contain around 100,000 copies of the genes
coding for 18S, 28S and 5.8S ribosomal RNAs. It doesn’t, however. Apparently
the load of carrying large numbers of these much bigger genes would be too
great. Instead, Xenopus generates the
required number of DNA templates for the other ribosomal RNAs by selective
replication of the 450 ribosomal RNA genes before the ribosomal RNA is to be
synthesized.
Once, a key to success in developmental biology was
finding an organism or cell-type devoted to the activity you wished to study.
In such cells the specialization in activity reduces the level of other and
inter-fering activities. This simplifies the experimenter’s measurements. Also,
the purification for biochemical study of the components involved is simpler
since they are present in sizeable concentrations in these cells. For this
reason Xenopus permitted careful
examination of several im-portant developmental biology questions before the
appearance of genetic engineering techniques.
The strategy of selective gene amplification used
by Xenopus on its ribosomal RNA genes
is not unique. Other organisms also utilize it for genes whose products are
needed in high level. In terminally differenti-ated tissues where no further
cell division occurs, the gene amplification can occur right on the chromosome
by the selective activity of a replication origin (Fig. 15.1). Since an egg
cell, however, must later divide, the mechanism of gene amplification must not
irreparably damage the genome. In Xenopus
a copy of the rRNA gene is replicated
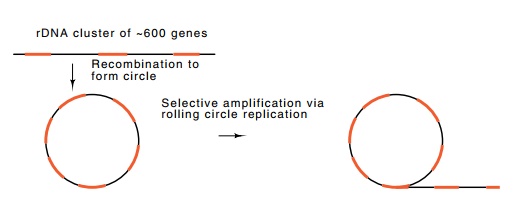
Figure
15.2 Selective replication of rRNA
genes yields as many as 4,000,000gene copies per cell.
free of the chromosome to generate high numbers of
templates neces - sary for the synthesis of the RNA (Fig. 15.2). As the last
components of a ribosome are synthesized, mature ribosomes form. Some begin
trans-lating maternal mRNA while others remain inactive until fertilization
triggers a more vigorous rate of protein synthesis.
Why the genes coding for 5S RNA cannot be amplified
along with the 18S, 28S, and 5.8S rRNA genes is not apparent. Xenopus, and most other organisms, have
gone to considerable trouble to separate the synthesis of 5S and the other
ribosomal RNAs. Even the RNA polymerases used are different: 5S uses RNA
polymerase III whereas 18S, 28S, and 5.8S use polymerase I.
As the oocyte matures, ribosome synthesis and most
other activity ceases until fertilization activates protein synthesis. Then,
after the developing embryo has reached the 4,000 cell stage, 5S ribosomal RNA
synthesis begins again. A small fraction of the RNA synthesized at this time is
the RNA species which was synthesized in the oocyte. Most, however, possesses a
slightly different sequence. It is called somatic 5S RNA. The oocyte 5S RNA
synthesis soon ceases altogether, leaving just the somatic 5S RNA synthesis.
The haploid genome of Xenopus
contains about 450 copies of the somatic variety of 5S gene in contrast to
about 100,000 copies of the variety expressed only in oocytes.
Study of 5S RNA synthesis is attractive because the
5S genes show developmental regulation in that the oocyte 5S genes are
expressed only in the developing oocyte whereas the somatic 5S genes are
expressed both in the oocyte and in the developing embryo. In addition to the
simple timing pattern of 5S RNAs, the product of the synthesis is simple. The
5S RNA is not processed by capping, polyadenylation, or splicing, nor is there
a tissue specificity in the expression. Hence this seems like an ideal material
for study of developmental questions.
Related Topics