Chapter: Genetics and Molecular Biology: Xenopus 5S RNA Synthesis
TFIIIA Binding to the Middle of its Gene as Well as to RNA
TFIIIA Binding to the Middle of its Gene as Well
as to RNA
Prokaryotic promoters are located upstream from
their genes. There-fore it was logical to look in front of 5S genes for the
sequences necessary for synthesis of 5S RNA. Finding these sequences could be
approached with the tools of molecular genetics. Nuclear extracts capable of
accu-rately synthesizing 5S RNA were used with pure DNA templates. A series of
templates could be generated with progressively larger deletions approaching,
and even entering the 5’ end of the 5S gene. After deletion of the natural
sequences, the plasmids were ligated closed and trans-formed into bacteria for
biological synthesis of large quantities of the DNA. The product of the
constructions was a substitution of plasmid sequence for what had been native Xenopus sequence ahead of the 5S gene
(Fig. 15.3).
The deletion experiments were remarkable. Deleting
all of the natural sequence ahead of the 5S sequence did not reduce synthesis
of the 5S RNA to less than 50% of normal. Even when the first 40 nucleotides of
the sequence were deleted, an RNA product of about the 120 nucleotide size of
5S RNA was synthesized. In this case the first 20 nucleotides synthesized were
specified by plasmid DNA sequence (Fig. 15.4). Only when the deletions extended
beyond about position 50 inside the 5S gene did they block synthesis.
The location of the downstream edge of the
sequences necessary for 5S RNA synthesis is also of interest. The experiments
to determine this position required a technical trick. In the experiments
probing for the
Figure
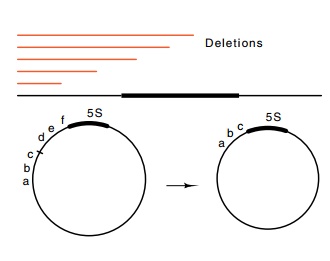
15.3 Deletions upstreamof a gene do
not eliminate DNA, they substitute new sequences like abc for the deleted DNA def.
Figure
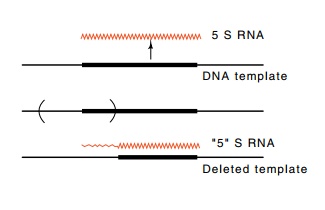
15.4 Deletion of partof the 5S gene and
all upstream sequences still yields transcrip-tion of the gene. Transcription
begins from approximately the same location as that used when the normal
sequence is present.
sequences necessary for transcription initiation,
the natural termina-tion sequence of the 5S gene remained intact. Therefore,
whatever the sequence of the RNA at its 5’ end, transcription still terminated
normally and yielded an RNA molecule of about 120 nucleotides. The presence of
this RNA could be determined simply by arranging that the RNA synthesized in vitro be radioactive and then running
the resultant products on an acrylamide gel. When 5S RNA had been synthesized
in a reaction, it formed a prominent band in the gel. Such a simple assay
technique could not work if the sequences at the 3’ end of the gene that are
required for termination were deleted.
Hybridization assays could be devised to look for
RNA sequences from the remaining 5’ end of the deleted 5S gene. These assays
however, would also detect any readthrough transcription resulting from
incor-rect initiation events occurring upstream of the 5S gene (Fig. 15.5). A
better assay for 5S RNA synthesis was to force the 5S RNA to terminate
prematurely, before it reached sequences that might be altered due to deletion
of the 3’ end of the gene. These terminations could be generated by introducing
a transcription terminator in the region between the start site of
transcription and the beginning of the sequence already deter-mined to be
required for transcription (Fig. 15.6). A simpler approach,
Figure
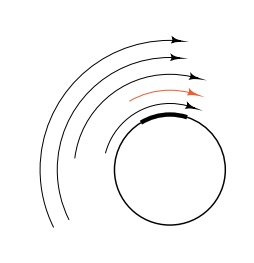
15.5 Mere total RNA in-cluding 5S
sequences is not a good assay for activity of specific tran-scription of the 5S
RNA gene due to nonspecific transcription which begins upstream of the gene.
Figure
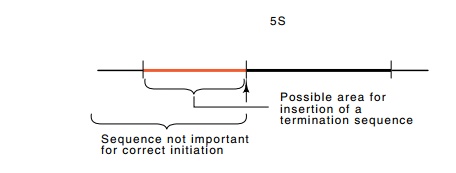
15.6 A region in which a transcription
terminator could be placed forassay of specific transcription of the 5S gene.
but one based on this basic idea, was taken. This
used the same trick as is used in DNA sequencing. In the triphosphates used for
transcription was included a low concentration of a 3’-dideoxy-triphosphate.
When a molecule of this analog was incorporated into the growing RNA chain, no
further elongation was possible. At appropriately low concentrations of a
3’deoxy-triphosphate, a characteristic spectrum of sizes of prema-turely
terminated RNA molecules results (Fig. 15.7). Transcription initiating at other
points on the chromosome generate other spectra of prematurely terminated
molecules. Despite the presence of these groups of RNA molecules, the spectrum
indicative of initiation from the 5S RNA promoter could be identified and
therefore the activity of this promoter could easily be assayed.
The experiments probing for the location of the 3’
end of the se-quences necessary for normal initiation of the 5S gene were
successful. The downstream edge of the sequences required was between positions
80 and 83. That is, within the gene, between positions 50 and 80 lies a stretch
of 30 nucleotides which are required for correct synthesis of the 5S RNA.
Does TFIIIA bind within the interior of the 5S
gene? Indeed it does. DNAse footprinting experiments show that it protects the
region from about +50 to +80. Similarly, pre-ethylation experiments in which
those phosphates are determined whose ethylation prevents or substantially
Figure
15.7 A spectrum of specific RNA sizes
generated by transcriptionbeginning at the 5’ end of the 5S RNA gene and
terminating at particular nucleotides.
weakens binding of the protein also revealed the
binding site of the protein to be entirely within the gene. These deletion and
biochemical experiments show that TFIIIA determines where RNA polymerase III
initiates on the 5S gene.
The TFIIIA protein revealed an additional
unexpected property. It was noticed that an excess of 5S RNA in the in vitro transcription reactions
inhibited transcription of the 5S gene. This type of result suggests that the
RNA inhibits RNA polymerase. This possibility could be simply tested since RNA
polymerase III also transcribes tRNA genes. If the 5S RNA inhibited RNA
polymerase, it should block synthesis of both 5S RNA and tRNA. It did not. It
blocked only 5S RNA synthesis. Therefore, 5S RNA must bind TFIIIA, thereby
titrating TFIIIA from the DNA. Indeed, this is true and the dual binding
ability of TFIIIA appears to be of biological importance as one of the proteins
in the 7S complex of 5S RNA is TFIIIA.
Related Topics