Chapter: Genetics and Molecular Biology: Regulation of Mating Type in Yeast
DNA Cleavage at the MAT Locus
DNA Cleavage at the MAT Locus
A key step in the shifting of mating type is the
transfer of the genetic information from HML
and HMR to MAT. Apparently the transfer is initiated by cleaving the DNA at
the MAT locus. A specific
endonuclease, much like a restriction enzyme, could generate a double-stranded
break in DNA at the MAT locus. How
could such an enzyme be sought? Restriction enzymes can be detected by their
cleavage of DNA, such as lambda phage or plasmid DNA. If the mating-type
endonuclease is specific, then only DNA carrying the MAT locus and possibly the HML
and HMR loci would be substrates.
Also, the enzyme generating the cleavages should be found in haploid HO yeast but probably would not be found
in ho or in diploid yeast
since these latter types do not switch mating type.
A
particularly sensitive assay was used to search for the predicted cleavage
(Fig. 16.14). A plasmid containing the mating-type region was cut and
end-labeled with 32PO4. Incubation of this DNA with
extracts prepared from various yeast strains should cut the DNA somewhere
within the MAT locus. Such cleavage
would generate a smaller radioac-tive fragment which could be detected on electrophoresis
of the diges-tion mixture and autoradiography. As expected, HO haploids contain a cleaving activity,
and, furthermore, this activity is present at only one part of the cell cycle,
just before the period of DNA synthesis. Using this cleavage assay, the enzyme
has been partially purified, and biochemical studies on it are now possible.
Figure 16.14 A sensitive scheme for detecting an endonuclease that cleavesspecifically in MAT sequences.
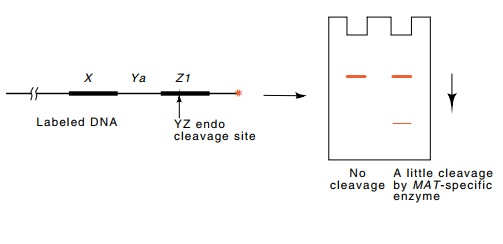
The site of DNA cleavage lies just within the MAT locus. Some of the mutations that act in cis to prevent the normal high frequency of mating-type conversion do not generate the
double-stranded break invivo. Most
likely they prevent binding or cleavage by the HO nuclease,and, as expected,
they prevent cleavage in vitro. As is
typical for most restriction enzyme cleavage sites, these changes lie within 10
base pairs of the cleavage site.
The HO endonuclease is particularly unstable.
Presumably this re-sults from the requirement that any cleavage that is to
generate a mating-type switch must be made in a narrow window of time. The
regulation of the synthesis of HO messenger RNA also must be tight. Apparently
a sizeable number of proteins are involved in this regulation. A region of DNA
preceding the HO gene of over a
thousand nucleotides and about fifteen different proteins seem to be involved.
Figure 16.15 The pattern ofmating type switching in HO S.cerevisiae. The star indicatesthat the mother cell will switch mating type in the next genera-tion. Red indicates a switch.
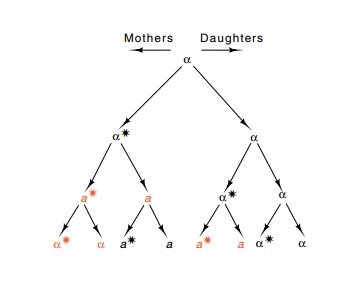
In HO cells the pattern of mating-type conversion
is fixed. Only in a cell that has produced at least one daughter, called a
mother cell, does a mating-type conversion occur (Fig. 16.15). At this time
both the mother and daughter convert. The complex set of proteins that regulate
HO protein expression also compute this cell lineage and permit expres-sion of
the HO gene only in the mother and
not in first-born daughters.
Related Topics