Chapter: 11th Biochemistry : Chapter 3 : Proteins
Proteins and their structure
Proteins and their structure
Proteins
are made up of the 20 different amino acids. These amino acids are joined
together by a covalent linkage commonly known as a peptide bond. The linear
sequence of these linked amino acids is specific for a protein. The amino acid
sequence contains necessary information for that protein to fold into a unique
three dimensional structure and correspondingly a unique
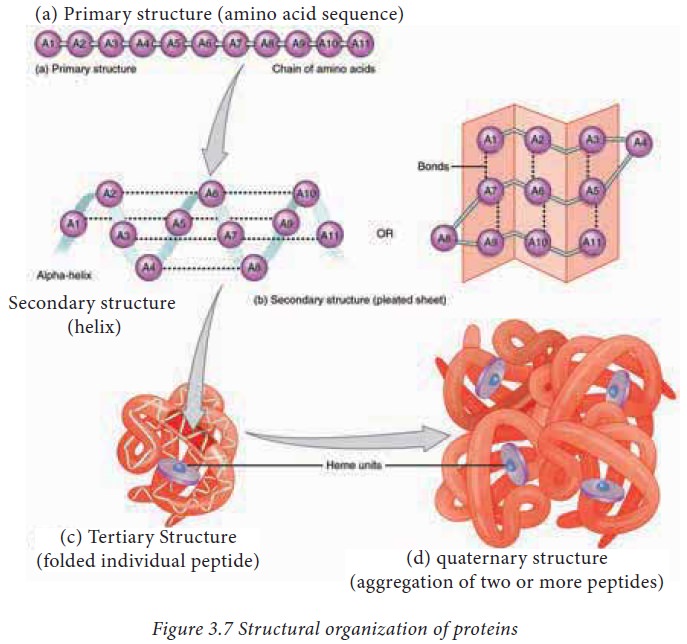
function. The structure of proteins can be best understood by considering them
in four hierarchical levels as described in figure 3.7
1. The primary structure of proteins
The
amino acid sequence of a protein is known as its primary structure. Knowing the
primary structure for a protein is important because even small changes (due to
mutations) in the primary structure can lead to improper folding and hence
impairment or complete loss of function.
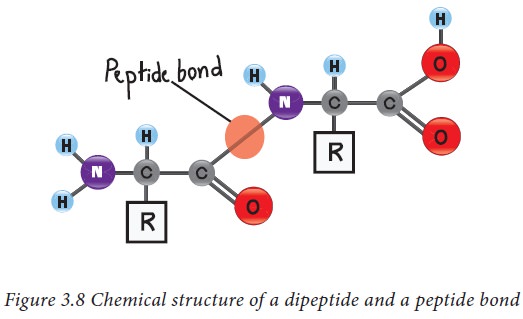
Peptide bonds
The amino acids in protein are
covalently linked together to form peptide bonds. Peptide bonds are amide
linkages between the α carboxyl group of one amino acid and the amino group of another amino acid. For example,
serine and alanine can form a peptide called serylalanine as described in
figure 3.8. Since two amino acids are joined together, this molecule is known
as a dipeptide. If many amino acids are joined together in the same way to form
a single chain, such a chain is known as a polypeptide. The atoms excluding the
side chains of amino acids in a polypeptide are known together as the back bone
or main chain of the polypeptide.
Peptide bonds have some important
properties, which are
a. Peptide bonds are generally in trans conformation. However in rare conditions peptide bonds formed
by proline can adopt a cis
conformation.
b. Peptide bonds have a partial double bond character, which gives
them a planar nature and hence cannot be rotated.
c.
Since peptide bonds
are amide linkages the –C=O and –NH groups cannot donate or accept protons and
are uncharged. The net charge of a polypeptide can come only from the N
terminus amino group, C terminus carboxyl group and the side chains of the
amino acids.
d. Despite not being ionisable, the –C=O and –NH groups of peptide
bonds are polar and can involve in the formation of hydrogen bonds. This
property is important for the formation of secondary structures of proteins.
2. Secondary structure of proteins
The back
bone of a polypeptide forms regular structural arrangements by making hydrogen
bonds with its neighbouring amino acids. As a rule, these hydrogen bonds are
always between the main chain –NH group and –C=O group. There are three main
types of secondary structures present in proteins namely α Helix, β sheet and β
turn.
Hydrogen Bonds
Hydrogen
bonds are weak electrostatic interactions between an electro negative atom and
a hydrogen which is covalently linked to another electro negative atom.
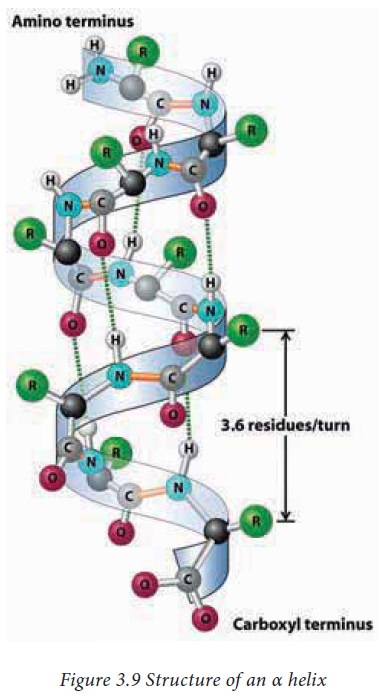
α Helix
It is a spiral (helical) structure of a tightly
packed and coiled main chain of a polypeptide with the side chain groups of amino
acids protruding outside. The helical structure is achieved by the formation of
hydrogen bonds between the –C=O of an nth amino acid with the –NH
group of n+4th amino acid. Each turn of an α helix contains 3.6
amino acids.The α helices are mostly right handed but there are rare instances
where left handed α helices are also present in proteins. The amino acid Proline
can produce a kink in an α helix as its secondary amino group is not
geometrically compatible inside an α helix.
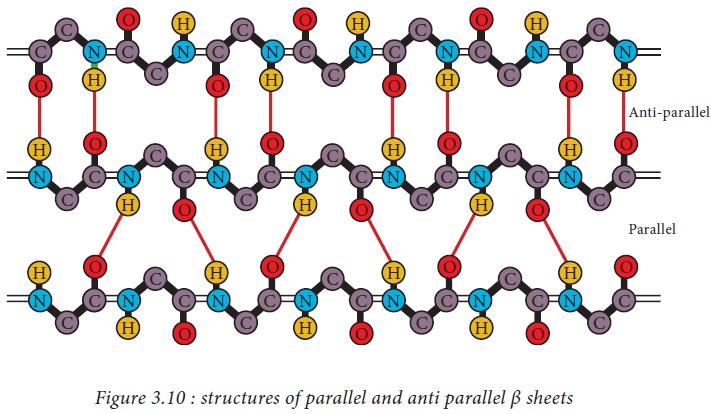
β pleated sheets
In a β
pleated sheet, two or more segments of a polypeptide chain line up next to each
other forming a sheet like structure held together by hydrogen bonds. The
strands of a β pleated sheet may be parallel where the N- and C- termini of the
strands match up or antiparallel where the N-terminus of one strand is
positioned next to the C-terminus of the other.
β turns
These
are secondary structural elements with four amino acids that can reverse (turn)
the direction of a polypeptide and thus help the polypeptide to form a globular
shape. They are mostly found on the surface of proteins. The amino acids
proline and glycine are more frequently found in β turns. They are also mostly
found to connect two different α helices or β strands to form super secondary
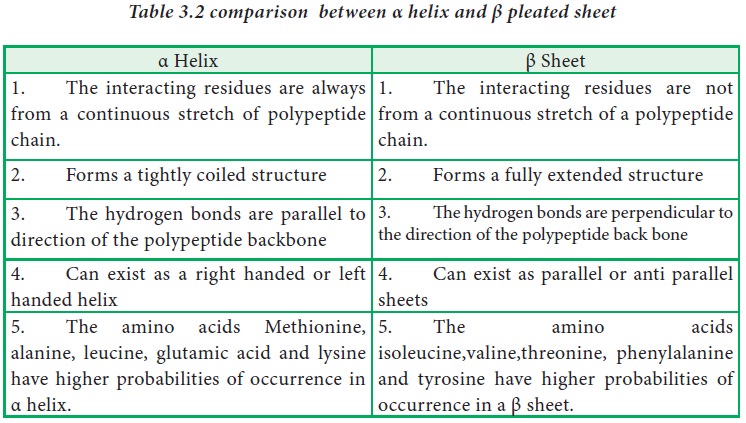
structure motifs such as helix-turn helix, beta meander, beta barrel etc.
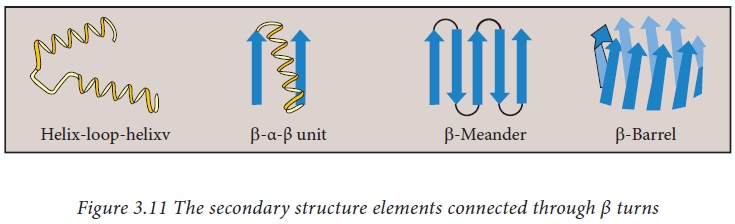
3. Tertiary structure
The
polypeptide folds in such a way that the secondary structure elements are
packed compactly to form an overall three-dimensional structure called its tertiary structure. The tertiary structure is stabilized mainly by the
interactions between the R groups
(side chains) of the amino acids.
The
interactions that contribute to tertiary structure are hydrogen bonds, ionic
interactions, dipole-dipole interactions and Vander Waals Forces. The above
mentioned interactions are also known as non bonded interactions.
The side
chains with like charges such as Lys and Arg repel one another, while those
with opposite charges such as Lys and Asp can form an ionic interaction.
Similarly, polar R groups can form hydrogen bonds and other dipole-dipole
interactions.
The
amino acids with non polar, hydrophobic R groups cluster together on the inside
of the protein through hydrophobic interactions. This cluster is also known as
the hydrophobic core and it is an important feature of globular proteins.
Similarly, the hydrophilic amino acids, (i.e.) the amino acids with side chains
containing charged groups are present on the surface of globular proteins to
interact with surrounding water molecules.
The
sulphur containing side chains of two cysteine residues can form a covalent
bond known as a disulfide bond. The
disulfide bonds help to bring together two different parts of
the same polypeptide or two different polypeptides together and are the only
covalent interactions involved in the formation of tertiary structure.
4. Quaternary structure of proteins
Proteins that are made up of a single
polypeptide chain have only three levels of structure. Some proteins are made
up of more than one polypeptide chain. In such cases the tertiary structures
formed by each of those polypeptide chains come together to form a quaternary
structure. These individual polypeptide chains are also known as subunits.
Hemoglobin, a protein which carries oxygen in blood is made up of four
subunits. Similarly, DNA polymerase, an enzyme which synthesizes new strands of
DNA is composed of ten subunits. The same types of interactions that contribute
to tertiary structure are also involved in stabilization of the quaternary
structure.
Related Topics