Chapter: Biotechnology Applying the Genetic Revolution: Basics of biotechnology
Packaging of Nucleic Acids
PACKAGING
OF NUCLEIC ACIDS
Bacteria have just a few
thousand genes, each approximately 1000 nucleotides long. These are carried on
a chromosome that is a single giant circular molecule of DNA. A single DNA
double helix with this many genes is about 1000 times too long to fit inside a
bacterial cell without being condensed somehow in order to take up less space.
In bacteria, the double helix
undergoes supercoiling to condense
it. Supercoiling is induced by the enzyme DNA
gyrase, which twists the DNA in a left-handed direction so that about 200
nucleotides are found in one supercoil. The twisting causes the DNA to
condense. Extra supercoils are removed by topoisomerase
I. The supercoiled DNA forms loops that connect to a protein scaffold (see
Fig. 1.4).
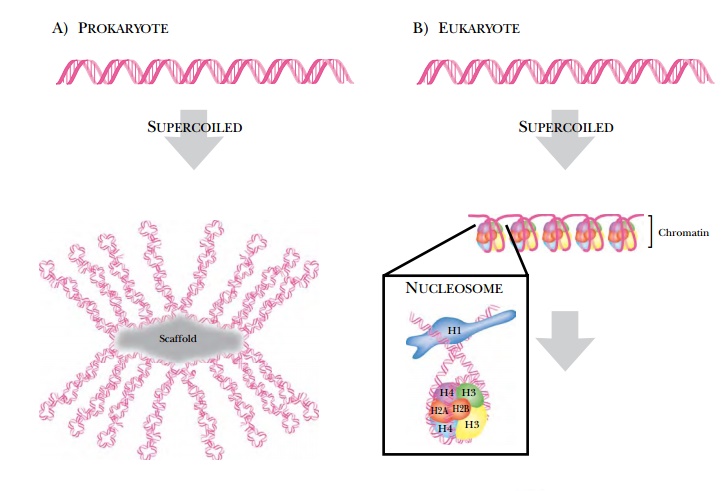
In humans and plants, vastly more DNA must be packaged, so just adding supercoils is not sufficient. Eukaryotic DNA is wound around proteins called histones first. Histones have a positive charge to them, and this neutralizes the negatively charged phosphate backbone. DNA plus histones looks like beads on a string and is called chromatin. Each bead or nucleosome has about 200 base pairs of DNA and nine histones, two H2A, two H2B, two H3, two H4, and one H1. All the histones form the “bead” except for H1, which connects the beads by holding the DNA in the linker region. The histones are highly conserved proteins that are found in all eukaryotes and, in simplified form, in archaebacteria. Histone tails stick out from the nucleosome and are important in regulation. In regions of DNA that are expressed, the histones are loose, allowing regulatory proteins and enzymes access to the DNA. In regions that are not expressed, the histones are condensed, preventing other proteins from accessing the DNA (this structure is called heterochromatin).
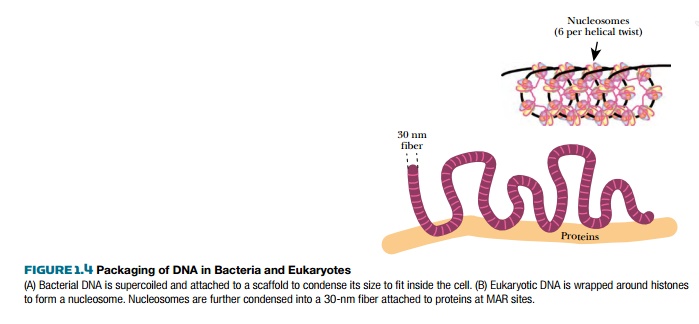
Chromatin is not condensed enough to fit the entire eukaryotic DNA genome into the nucleus. It is coiled into a helical structure, the 30-nanometer fiber, which has about six nucleosomes per turn. These fibers loop back and forth, and the ends of the loops are attached to a protein scaffold or chromosome axis. These attachments occur at matrix attachment regions (MAR) and are mediated by MAR proteins. These sites are 200–1000 base pairs in length and have 70% A/T. The structure of A/T-rich DNA is slightly bent, and these bends promote the connection between proteins in the matrix and the DNA. Often, enhancer and regulatory elements are also found at these regions, suggesting that the structure here may favor the binding of protein activators or repressors. This structure refers to chromosomes during normal cellular growth. When a eukaryotic chromosome readies for mitosis and cell division, it condenses even more. The nature of this condensation is still uncertain.
DNA must be condensed by supercoiling and
wrapping around nucleosomes to form chromatin, and finally attached to protein
scaffolds in order to fit into the nucleus.
Related Topics