Chapter: Plant Biochemistry: Nitrate assimilation is essential for the synthesis of organic matter
Nitrate assimilation is strictly controlled
Nitrate assimilation is strictly controlled
During photosynthesis, CO2 assimilation and nitrate assimilation have to be matched to each other. Nitrate assimilation can progress only when CO2 assimilation provides the carbon skeletons for the amino acids. Moreover, nitrate assimilation must be regulated in such a way that the production of amino acids does not exceed the demand. Finally, it is important that nitrate reduction does not proceed faster than nitrite reduction, to prevent the accumulation of toxic levels of nitrite. For example, dan-gerous levels of nitrite may accumulate under anaerobic soil conditions in the case of excessive moisture. Flooded roots are able to release nitrite into the soil water, avoiding the buildup of toxic levels of nitrite. This escape route, however, does not function in leaves, and there the strict control of nitrate reduction is especially important.
The NADH required for nitrate reduction in the cytosol can also be provided during darkness (e.g., by glycolytic degradation of glucose). The reduction of nitrite and fixation of NH4+ in the chloroplasts depends largely on photosynthesis providing reducing equivalents and ATP, whereas the oxidative pentose phosphate pathway can offer only limited amounts of reducing equivalents in the dark. Therefore, during darkness nitrate reduction in the leaves has to be slowed down or even switched off to prevent an accumulation of nitrite. This illustrates how essential it is for a plant to regulate the activity of nitrate reductase, which is the entrance step of nitrate assimilation.
The synthesis of the nitrate reductase protein is regulated at the level of gene expression
Nitrate reductase is an exceptionally short-lived protein. Its half-life time is only a few hours. The rate of de novo synthesis of this enzyme is very high. Thus, by regulating its synthesis, the activity of nitrate reductase in the tis-sue can be altered within hours.
Various factors control the synthesis of the enzyme at the level of gene expression. Nitrate and light stimulate the enzyme synthesis. Part of the light effect is caused by carbohydrates generated by photosynthesis. The synthesis of the nitrate reductase protein is stimulated by glucose and other carbohydrates generated by photosynthesis, and is inhibited by NH4+ , glutamine and other amino acids (Fig. 10.9). Sensors seem to be present in the cell that adjust via regulation of gene expression the capacity of nitrate reductase both to the demand for amino acids and to the supply of carbon skeletons from CO2 assimilation for its synthesis.
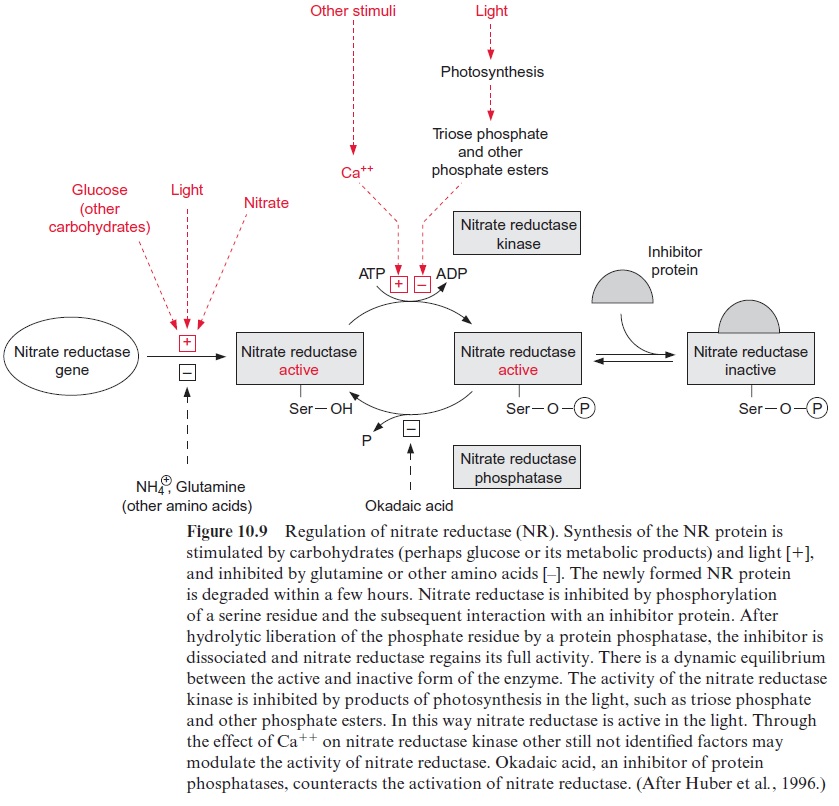
Nitrate reductase is also regulated by reversible covalent modification
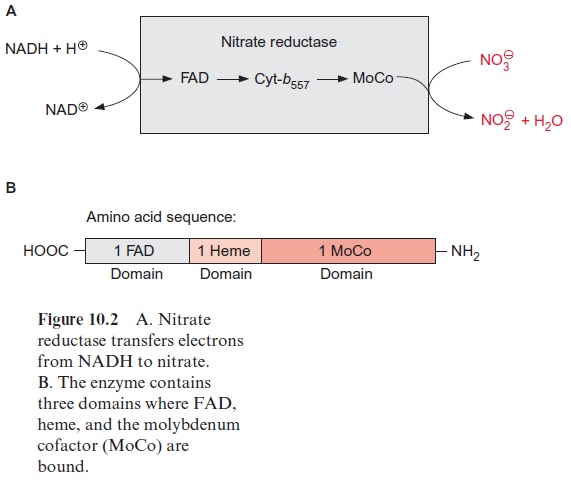
The regulation of de novo synthesis of nitrate reductase (NR) allows regula-tion of the enzyme activity within a time span of hours. This would not be sufficient to prevent an acute accumulation of nitrite in the plants during darkening or sudden shading of the plant. Rapid inactivation within min-utes of nitrate reductase occurs viaphosphorylation of the nitrate reductase protein (Fig. 10.9). Upon darkening, a serine residue, which is located in the nitrate reductase protein between the heme and the MoCo domain, is phosphorylated by a protein kinase termed nitrate reductase kinase. This protein kinase is inhibited by the photosynthesis product triose phosphate and other phosphate esters and is stimulated by Ca++ ions, a messenger compound of many signal transduction chains. The phos-phorylated nitrate reductase binds aninhibitor protein, which interrupts the electron transport between cytochrome-b557 and the MoCo domain (Fig. 10.2). The nitrate reductase phosphatase hydrolyzes the enzyme’s serine phosphate and this causes the inhibitor protein to be released from the enzyme and thus nitrate reductase is restored to an activated state. Okadaic acidinhibits nitrate reductase phosphatase and in this way also inhibits the reactivation of nitrate reductase. Since the phosphorylation of the serine residue and the binding of the inhibitor protein are reversible, there is a dynamic equilibrium between the active and inactive form of the nitrate reductase. The inhibition of nitrate reductase kinase by triose phos-phate and other phosphate esters ensures that nitrate reductase is active only when CO2 fixation is operating for delivery of the carbon skeletons for amino acid synthesis, which is discussed in the next section.
14-3-3 proteins are important metabolic regulators
It was discovered that the nitrate reductase inhibitor protein belongs to a family of highly conserved regulatory proteins called 14-3-3 proteins, which are widely spread throughout the animal and plant worlds. 14-3-3 proteins bind to a specific binding site of the target protein with six amino acids (Arg-X-X-SerP/ThrP-X-Pro), which contain a serine or threonine phos-phate in position 4. The importance of these latter amino acids for nitrate reductase was verified in an experiment, in which the serine in the 14-3-3 protein binding site of nitrate reductase was exchanged for alanine via site directed mutagenesis; the altered nitrate reductase was no longer inacti-vated by phosphorylation. 14-3-3 proteins bind to a variety of proteins and change their activity. They form a large family of multifunctional regulatory proteins, many isoforms of which occur in a single plant. Thus 14-3-3 pro-teins regulate in plants the activity of the H+ -P-ATPase of the plasma membrane. 14-3-3 proteins regulate the function of transcription factors and protein transport into chloroplasts. There are indications that 14-3-3 proteins are involved in the regulation of signal transduction as they bind to various protein kinases and play a role in defense processes against biotic and abiotic stress. The elucidation of these various functions of 14.3.3 proteins is at present a very hot topic in research.
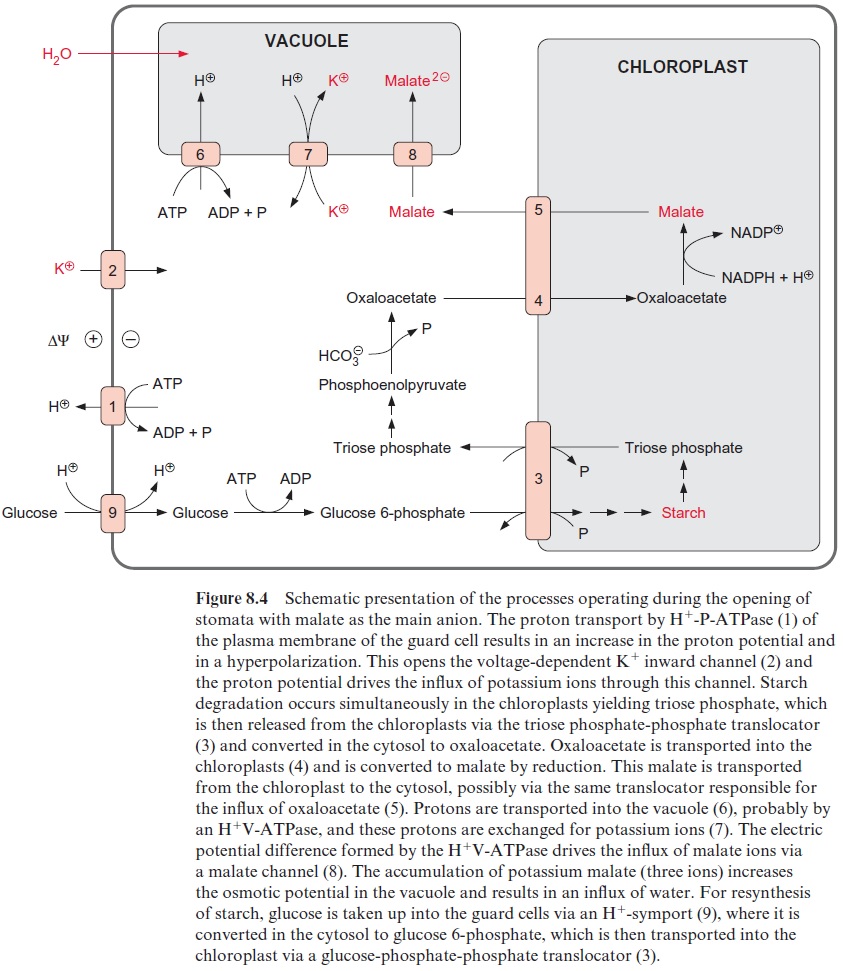
This important function of the 14-3-3 proteins in metabolic regulation is exploited by the pathogenic fungus Fusicoccum amygdalis to attack plants. This fungus forms the compound fusicoccin, which binds specifically to the 14-3-3 protein binding sites of various proteins and thus cancels the regula-tory function of 14-3-3 proteins. In this way fusicoccin disrupts the metab-olism to such an extent that the plant finally dies. This attack proceeds in a subtle way. When F. amygdalis infects peach or almond trees, at first only a few leaves are affected. In these leaves the fungus excretes fusicoccin into the apoplasts, from which it is spread via the transpiration stream through the other parts of the plant. Finally, fusicoccin arrives in the guard cells; where it causes an irreversible transformation of the H+ -P-ATPase into the active form resulting in continuously opened stomata (Fig. 8.4). This leads to a very high loss of water; consequently, the leaves wilt, the tree dies, which ultimately is the nutrient source of the fungus.
There are great similarities between the regulation of nitrate reductase and sucrose phosphate synthase
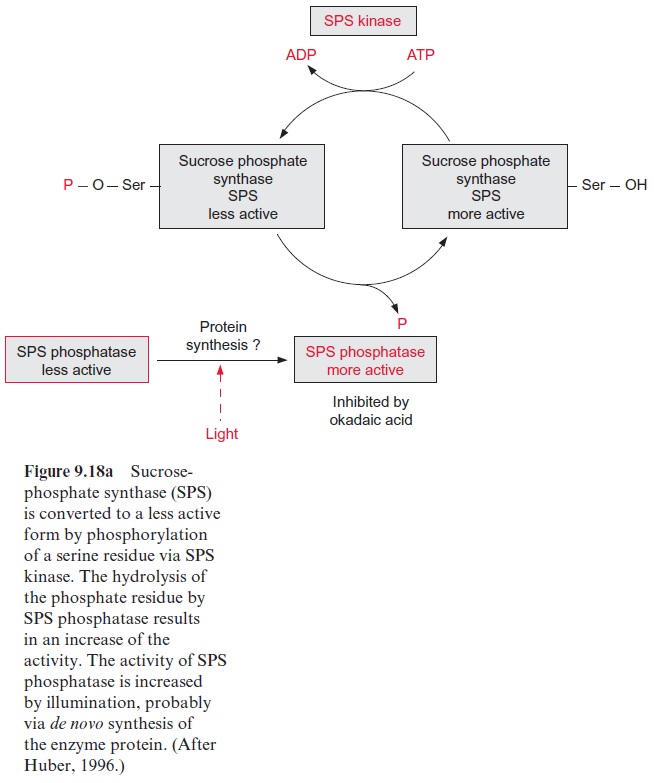
The regulation principle of nitrate reduction by phosphorylation of serine residues by special protein kinases and protein phosphatases is remarkably similar to the regulation of sucrose phosphate synthase discussed (Fig. 9.18). Upon darkening, both enzymes are inactivated by phosphor-ylation, which in the case of nitrate reductase also requires a binding of an inhibitor protein. Both enzymes are reactivated by protein phosphatases, which are inhibited by okadaic acid. Also, sucrose phosphate synthase has a binding site for 14-3-3 proteins, but its significance for regulation is not yet clear. Although many details are still not known, it is obvious that the basic mechanisms for the rapid light regulation of sucrose phosphate synthase and nitrate reductase are similar.
Related Topics