Chapter: Plant Biochemistry: Biotechnology alters plants to meet requirements of agriculture, nutrition and industry
Genes can be turned off via plant transformation
Genes can be turned off via plant transformation
In addition to producing transgenic plants with new properties by the transfer and expression of foreign genes, it is of great interest to decrease or even eliminate certain properties by inhibiting the expression of the corre-sponding gene, for instance the activity of a translocator or of an enzyme. Turning off a gene is also an important way to identify its function by investigating the consequences of the missing gene on plant metabolism.
Homologous recombinations frequently occur in prokaryotic genomes (including the mitochondrial and plastid genomes) as well as in the nuclear genome of animals. This property is utilized in animals to eliminate the function of a defined gene by generating a so-called “knock-out” mutant. To eliminate the function of a gene of interest, a section of this knock-out mutant has to be isolated and cloned. Thereafter its sequence is altered by genetic techniques, and then subsequently reintroduced into the nucleus, where it can replace the unaltered gene by homologous recombination and the function of the gene is eliminated in the organism. In plants, however, this technique is usually not applicable, since in the nuclear genome of plants homologous recombinations are rare. Therefore alternative meth-ods are required in plants. A decrease in the expression of a gene can be achieved by inactivating the encoding mRNA by the synthesis of a com-plementary RNA, called antisense RNA (Fig. 22.20). Normally, mRNA occurs as a single strand, but in the presence of an antisense RNA strand, it can form a double-stranded RNA. Double-stranded RNA is unstable and degrades rapidly by ribonucleases. Therefore mRNA loses the ability to encode protein synthesis when the corresponding antisense RNA is present.
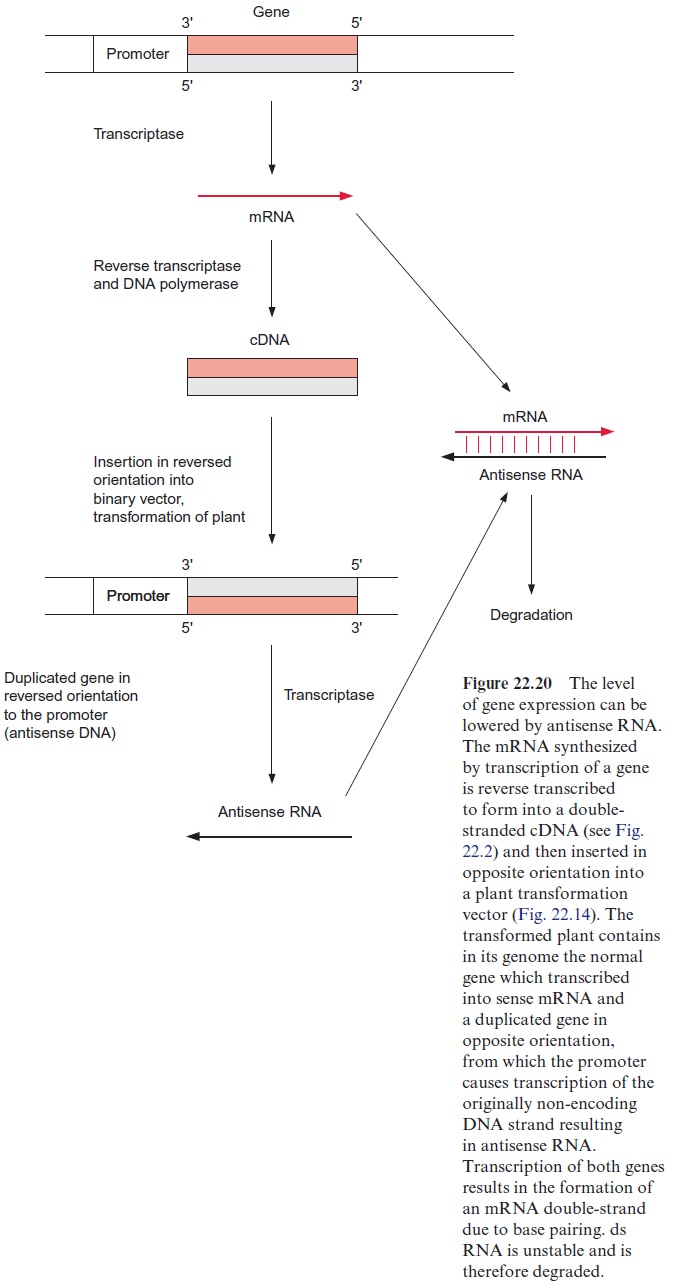
To synthesize the antisense RNA, the corresponding gene is first isolated according to the methods already described and then inserted as cDNA in reverse orientation in a vector, able to transform the plant of interest. As the orientation is reversed in this inserted gene (the promoter is positioned at the wrong side), the RNA polymerase transcribes the antisense RNA. This antisense technique has become a very important tool in plant genetic engineering particularly when decreased expression levels of a defined gene are required. Unfortunately this method does not always lead to the desired results. In many cases, it turned out to be impossible to decrease the syn-thesis of a protein (e.g., an enzyme or a translocator) by more than 90%. In some cases the activity of the remaining protein could be even enhanced by enzyme regulation, counteracting the intended elimination of gene func-tion. The antisense technique is particularly problematic when the corre-sponding gene is part of a multi-gene family.
A very effective new technique for eliminating the expression of a defined gene is the RNAi-technique (RNA-mediated-interference) . In this method, a double-stranded RNA (dsRNA) is prepared from a small RNA section (up to 30 nucleotides) of the mRNA to be inac-tivated. It was found that the presence of such dsRNA inhibits very effec-tively the translation of the respective mRNA and causes it to be degraded by the cell’s nucleases (RNase II). Therefore it is possible to eliminate the expression of a certain gene by transforming a plant with DNA encoding both strands of the dsRNA. Since the RNAi technique appears to have advantages over the antisense technique it is widely used.
For many plants, the transformation by A. tumefaciens is a simple way to eliminate gene functions at random. Plants take up the T-DNA of the Ti-plasmids and integrate them randomly into the genome. When the T-DNA is integrated into a gene, this gene is mutated (T-DNA insertion mutant) and therefore loses its function. The mutant can be subse-quently identified by a probe for the employed T-DNA.
Related Topics