Chapter: Health Management in Aquaculture: Immunological and molecular biology techniques in disease diagnosis
Gene probe assays - Aquaculture Molecular biology techniques in disease diagnosis
Gene probe assays
The power of DNA diagnostics is a consequence of two facts: (1) nucleic acidscan be rapidly and sensitively measured, and (2) the sequence of nucleotides in a given DNA molecule is so specific that hybridization analyses can be used for reliable clinical diagnoses.
One of the most powerful analytical tools available is nucleic acid hybridiza-tion. Instead of detecting a whole organism or its products, hybridization de-tects the presence or absence of specific DNA sequences associated with a spe-cific organism. To identify a microorganism through DNA analysis, you must have available nucleic acid probe to that microorganism, a single strand of DNA containing sequences unique to the organism. The unlabeled strand in the sample being analyzed that is homologous to the probe is called the target. If a microorganism in a specimen contains DNA sequences complementary to the probe, the two sequences can hybridize forming a double stranded mol-ecule. To detect that a reaction has occurred, the probe is labeled with a re-porter molecule, either a radioisotope, an enzyme, or a fluorescent compound that can be measured in small amounts following hybridization. Depending on the reporter used (radioisotopes are the most sensitive), as little as 0.25 mg of DNA per sample can be detected.
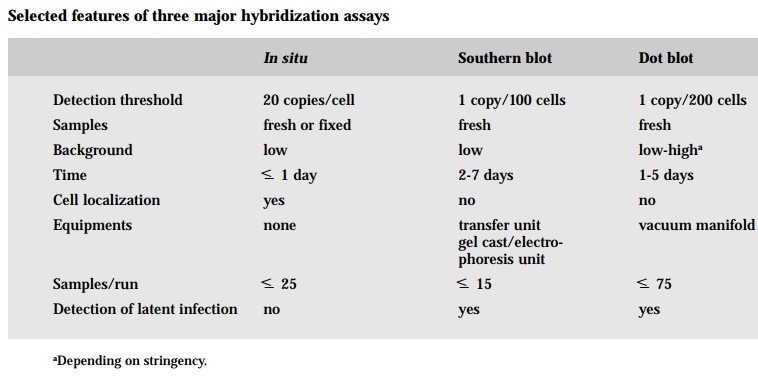
Several techniques have evolved based on the ability of a labeled probe to bind to and thus permit the detection of the target nucleic acid sequence of interest. One approach is to extract the DNA, both target and non-target, from a sample and bind it to a filter where it can be hybridized with the labeled probe. This is called filter hybridization. Often times the sample DNA is directly placed on the filter with the aid of a vacuum manifold, which has slot-like spaces for each sample, hence the term slot blot or dot blot hybridization. Dot blotting is rapid, simple technique for the quantification of RNA or DNA target sequences without prior electrophoretic separation. This method differs only in the shape of the immobilized nucleic acid spot deposited on the membrane. Nucleic acid is applied to a dry nitrocellulose filter and allow to dry. The resulting “dots” are variable in size, making accurate estimates of target sequence concentra-tion difficult. Alternatively, the sample DNA may first be separated according to size and configuration by electrophoresis on a gel and then transferred to a filter - this is termed Southern blot hybridization. As can DNA fragments, RNA molecules can be separated on the basis of size by gel electrophoresis and can be immobilized on membranes by a process referred to as Northern blot hy-bridization. Detection of target sequences in Southern, Northern and dot blots is carried out under essentially identical conditions. Three basic processes are involved: (1) prehybridization, which saturates nonspecific DNA binding sites on the membrane with random DNA and polymers; (2) hybridization, during which specific labeled probes are annealed to target sequences; and (3) wash-ing, to remove unhybridized and imprecisely hybridized probe. In either of these techniques, the tissue must be destroyed thus precluding direct histologi-cal correlation.
In the other major strategy based on hybridization of a target and a probe, the target DNA is not extracted but rather kept in the intact cell where it may bind to the probe - this is the in situ hybridization. The principal advantage of insitu over other molecular techniques is it provides information about the loca-tion of the target nucleic acids within cells and/or tissues. Pathologists are able to look for specific nucleic acids and to study the cellular and tissue morphol-ogy of the sample. A misconception about in situ hybridization is that one needs to use radiolabeled probes in order to maximize its sensitivity however, recent and dramatic advances in nonisotopic labeling and, more importantly, detection systems has greatly enhanced the sensitivity using such common la-bels as biotin and digoxigenin. The most common problem encountered with the use of nonradioactive systems is background. Background may be defined as the presence of a hybridization signal with a specific probe in areas of the tissue where the signal should not be present. Background is often the result of nonspecific binding of the probe to nontarget molecules. Two ways to deal with background are to decrease the concentration of the probe and/or to in-crease the stringency of the post-hybridization wash. Another common prob-lem is poor tissue morphology due to overtreatment with the protease solution. Decreasing the time of digestion or the concentration of the protease tenfold will solve this problem. Another is occasionally tissue sections may fall off. The problem rests with incorrect silanization of the slides. The most obvious potential problem is the absence of a hybridization signal. In dealing with a negative signal, you have to check the proper fixative (no heavy metals or pi-cric acid), alkaline phosphatase conjugate, chromagen, denaturing tempera-ture (=95°C) and the tissue was deparaffinized. The target molecules are nucleic acids. Nucleic acids are complexed with proteins in the cell; when a tissue is embedded in a complex matrix, the nucleic acids are cross-linked to that matrix. Thus, major challenges of in situ hybridization are to make target nucleic acid available to the probe; and once proper hybrids are formed, to stabilized them without destroying the cell morphology.
An important difference between filter and in situ hybridization is the detec-tion threshold. Detection of a DNA sequence by in situhybridization implies a selective increase in its numbers due to, for example, oncogene amplification or viral proliferation. On the other hand, only one virus need to be present per every 100 cells for the Southern blot test to detect it, though the in situ test would be scored as negative. In situ is relatively insensitive test and is usually negative in situations such as occult or latent infection by a virus where the number of target DNA sequences per cell is low. Why is there such a disparity in detection threshold? The primary reason is the probe may find it more diffi-cult to find the target if it has to traverse the labyrinth of nuclear proteins and nucleic acids in in situ analysis relative to more “naked” DNA that has been attached to a filter in dot blot or Southern blot hybridization. Second, the extraction and purification of DNA characteristic of filter hybridization leads to a concentrating effect of rare nucleic acid sequences.
Nucleic acid probes offer many advantages over immunological assays. Nucleic acids are much more stable than proteins to high temperatures, high pH, organic solvents and other chemicals. In addition, nucleic acid probes are more defined entities than antibodies.
Gene probes are extensively used as diagnostic tools to detect white spot syn-drome virus (WSSV) (Chang et al., 1996; Chang et al., 1998), infectious hypo-dermal and hematopoietic necrosis virus (IHHNV) (Mari et al., 1993) and hepatopancreatic parvovirus (HPV) (Mari et al., 1995) of penaeid shrimp and viral hemorrhagic septicemia virus (VHSV) and infectious hematopoietic ne-crosis virus (IHNV) in fish (Ristow et al., 1991).
Related Topics