Chapter: Microbiology and Immunology: Virology, Virus: Orthomyxoviruses
Epidemiology - Influenza Viruses
Epidemiology
Influenza occurs worldwide.
◗ Geographical distribution
Influenza viruses cause epidemic, endemic, and pandemic influenza. Influenza A virus causes epidemics and occasionally pandemics; influenza B virus only causes epidemics; and influ-enza C viruses only cause minor respiratory illness and do not cause any epidemics. Influenza epidemics have been recorded throughout the history. The worst of these was the 1918 pan-demic, which caused about 20 million deaths worldwide and about 500,000 deaths in the United States. In temperate cli-mates, the epidemics typically occur in the winter and cause considerable morbidity in all age groups.
Influenza occurs throughout India. The worst, severe pandemic in the year 1918–1919 (Spanish flu) caused nearly 10 million deaths in India.
◗ Reservoir, source, and transmission of infection
Infected humans are the main reservoir of infections for influenza A virus. Respiratory secretions of infected persons are the important source of infection. The virus is excreted in respiratory secretions immediately before the onset of ill-ness and for 3–4 days thereafter. Wild aquatic birds are known reservoirs of influenza A. They secrete the viruses in their feces, which contaminates ponds and lakes. The virus is spread from person-to-person primarily by air-borne respiratory droplets released during the acts of sneezing and coughing.
Influenza B virus only causes epidemics. Infection is from humans-to-humans. No animal reservoir hosts are known.
Influenza C virus only causes minor respiratory illness and does not cause any epidemics.
◗ Influenza epidemics and pandemics
Influenza epidemics are of two types. Yearly epidemics are caused by both type A and type B viruses. The rare, severe influ-enza pandemics are always caused by type A virus. Antigenic shift and antigenic drift are the two different mechanisms responsible for producing the strains that cause these two types of epidemics.
Antigenic shift: A major change in one or both of the surfaceantigens, a change that yields an antigen showing no serologic relationship with the antigen of the strains prevailing at the time is called antigenic shift. Antigenic shift has been demon-strated in type A influenza virus only.
Influenza A virus causes epidemics and occasionally, pan-demics. Pandemics are caused by the virus strains undergoing antigenic shift. Antigenic shift variants appear less frequently, about every 10 or 11 years. It is demonstrated that pandemic strains are the recombinant strains, originated from some ani-mal or bird reservoir, either spreading to humans directly by host range mutation or as a result of a recombination between human and nonhuman strains. The pandemic strains also show the capability to spread rapidly among the population.
The completely novel antigens that appear during antigenic shift are acquired by genetic reassortment. The donor of the new antigens is probably an animal influenza virus. Type A viruses have been identified in pigs, horses, and birds, and animal influ-enza viruses possessing antigens closely related to those of human viruses. Fourteen distinct HA and nine NA antigens are known. Since continued surveillance of animal influenza viruses in recent years has failed to discover new antigens, these may represent the full variety of major influenza virus surface antigens (subtypes).
Antigenic drift: Repeated minor antigenic changes, on theother hand, generate strains that retain a degree of serologic relationship with the currently prevailing strain. This is called antigenic drift. The epidemics are caused by influenza A virus undergoing antigenic variations due to antigenic drift result-ing from mutations and selections. Antigenic drift variants occur very frequently, virtually every year. This is responsible for emergence of the strains that cause yearly influenza epidem-ics. When persons are reinfected with drift viruses, the serum antibody responses to the surface antigens that are shared with earlier strains to which the person has been exposed are fre-quently stronger and of greater avidity than are the responses to the new antigens. This phenomenon, which has been called “original antigenic sin” is sometimes useful in serologic diagnosis. Antigenic drift represents selection for naturally occurring vari-ants under the pressure of population immunity.
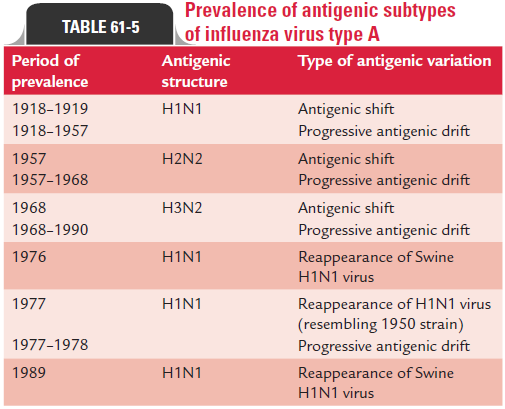
Influenza A is generally more pathogenic than influenza B. Influenza A is a zoonotic infection and more than 100 types of influenza A viruses have been described which infect birds, pigs, horses, dogs, and seals. The 1918 pandemic of influenza that resulted in millions of human deaths worldwide is believed to have originated from a virulent strain of H1N1 from pigs or birds (Table 61-5).
◗ H5N1 bird flu
The H5N1 flu, caused by an avian subtype influenza virus, which has been associated with bird flu in the domesti-cated birds, can be transmitted from birds to humans. The H5N1 was first described in Hong Kong in 1997. Human infections caused by this virus were established in only 18 individuals of which six died. Since then, sporadic cases of H5N1 infection continued to be described in southern China.
An epidemic of bird flu occurred in domesticated birds in Southeast Asia, primarily Vietnam, in January 2004. More than 240 human cases have been documented and more than 140 persons have died due to the poultry outbreaks and bird-to-human transmission. Most deaths have been reported in Vietnam and Indonesia. Sporadic outbreaks have continued to occur since the 2004 outbreak even outside Southeast Asia, including Turkey.
Till now, no conclusive evidences are available to show the human-to-human transmission of H5N1. However, scientists are concerned that a slight mutation could con-vert H5N1 to a strain that would be easily transferred from human-to-human. Such a strain, it is believed, could poten-tially spread rapidly and cause a catastrophic worldwide pan-demic. Hence, efforts are currently underway to develop an effective vaccine against H5N1. In addition, the number of drugs that are effective against influenza are being increas-ingly evaluated. Ribavirin has shown activity when tested in animal models.
Another avian subtype, H9N2, was described in two young children in March 1999. However, after that despite concern, no further outbreak of H9N2 infection has been documented. Experts are also concerned that a virulent strain of H9N2 influenza similar to H5N1 flu may mutate to allow human-to-human infection and that such a strain may possess the combination of transmissibility, infectivity, and lethality.
◗ H1N1
A novel H1N1 swine origin influenza virus (SOIV) H1N1 emerged in 2009 to produce the first human influenza pandemic of the twenty-first century. Within 1 year, this virus spread to 214 countries and caused more than 18,000 confirmed deaths worldwide. First described in April 2009, the novel swine origin influenza virus emerged due to a “triple-reassortment” of influenza viruses of avian, swine, and human origin. This led to the popular term “swine flu” being used for this virus. The 2009 pandemic H1N1 virus is composed of (a) PB2 and PA segments from North American avian viruses; (b) the PB1 segment of the human H3N2 viruses; (c) hemagglutinin (HA; of the H1 sub-type), nucleoprotein (NP) and NS segments derived from classical swine H1N1 viruses, and the neuraminidase (NA; of the N1 subtype), and (d) M segments of Eurasian “avian-like” swine viruses. Unlike previous strains of influenza virus, the novel swine origin influenza virus did not have a predilec-tion for populations more than 60 years of age. Rather, more number of younger people less than 40 years of age were found to be affected. A small percentage of those affected were found to develop pneumonia or acute respiratory dis-tress syndrome. The number of cases decreased greatly by May 2010, and the pandemic was officially declared to be at an end in August 2010. For diagnosis of H1N1 influenza, testing of samples like nasopharyngeal swabs or oropharyn-geal swabs is essential. Real time RT-PCR for viral nucleic acid is the recommended method for its diagnosis, and other rapid tests like antigen detection were not found to be useful.
The treatment for severe cases of H1N1 influenza is the neuraminidase inhibitor, oseltamivir.
Two types of influenza vaccines have been developed against H1N1 influenza—a trivalent inactivated vaccine and a live attenuated influenza vaccine, which can be administered intranasally.
Related Topics