Chapter: Pharmaceutical Biotechnology: Fundamentals and Applications : Oligonucleotides
Diagnostic Applications
DIAGNOSTIC APPLICATIONS
Apart from the direct application of ONs in medicine as therapeutics,
they also fulfill an increasingly important role as diagnostic agents. The
highly specific binding of complementary oligonucleotide sequences can be used
to detect gene expressionprofiles and mutations, whereas aptamers can be used
to detect presence of specific compounds. PCR amplification of specific nucleic
sequences can pro-vide information on the presence and abundance of this
particular sequence. For example, the Amplicor HIV-1 Monitor v1.5 assays
manufactured by Roche Diagnostics are currently approved for in vitro
diag-nostic use to determine viral load in blood samples.
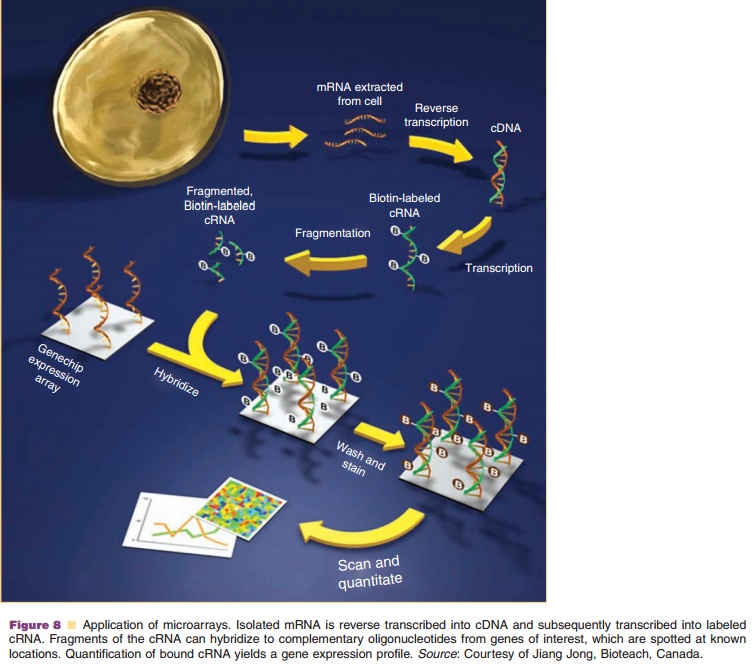
Other applications, like microarrays, do not focus on single genes but
provide an overview of the “transcriptome” (Fig. 8). A DNA microarray consists
of ONs of approximately 25 bases that are spotted on a chip in an orderly
arrangement, representing the genes of interest of an organism. Each ON is
spotted at a specific location on the array so that the location of each ON
with corresponding gene is known. Robotic spotters can currently place tens of
thousands of ONs accurately on one slide of a few square centimeters. Each spot
contains identical single-stranded ONs that are strongly attached to the slide
surface, allowing cellular DNA or RNA to be labeled and hybridized to the
complementary sequence on the array. By quantify-ing the binding of the labeled
DNA or RNA to the specific spots, the abundance of each species can be
determined and related to the corresponding gene.
In 2004, the FDA approved the first microarray AmpliChip CYP450 for
clinical use. The AmpliChip CYP450 provides complete coverage of the gene
variations, including duplications and deletion, of the cytochrome P450 enzymes
2D6 and 2C19. These genes are involved in the metabolism of approximately 25%
of all prescription drugs. It could be regarded as an important step towards
personalized medicine.
The inherent specificity and selectivity of aptamers makes these ONs
very useful to detect disease-associated molecules, in a similar manner as
antibody-based immunoassays. However, they are not in clinical use yet.
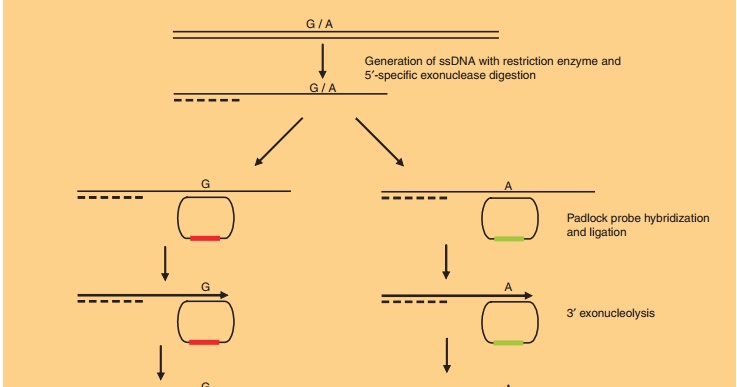
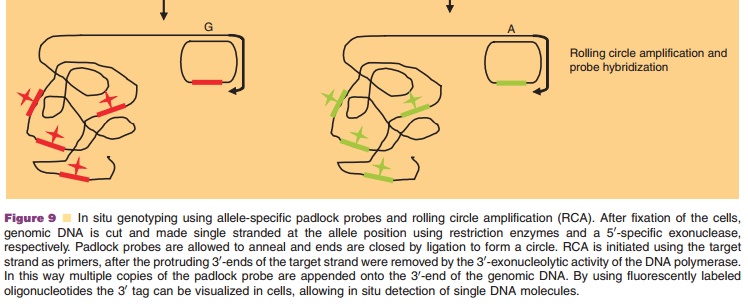
Padlock probes can also be used for diagnostic applications (Nilsson et
al., 1994). They consist of long ONs, whose ends are complementary to adjacent
target sequences. Upon hybridization, the ends of the ONs are brought together,
allowing ligation of the ON ends into a closed and intertwined circle that
cannot be replaced by the complementary DNA strand. This closed circle can then
be amplified by rolling circle amplification, a powerful and robust DNA
amplifica-tion method based on the mesophilic Phi29 DNA polymerase, allowing
amplification of the padlock signal to detectable levels (Fig. 9). Padlock
probes are more specific than conventional antisense ONs, as misannealing of
either one of the ends does not result in proper ligation of the ends, and thus
prevents circularization of the padlock probe. It has been used for multiplex
detection of pathogens in biological samples (Szemes et al., 2005), single
nucleotidepolymorphisms (Bakht and Qi, 2005) and miRNA (Jonstrup et al., 2006),
but also for in situ genotyping of individual DNA molecules (Larsson et al.,
2004).
Related Topics