Chapter: Medical Immunology: Major Histocompatibility Complex
Classification, Structure, and Detection of HLA Genes and Gene Products
CLASSIFICATION, STRUCTURE, AND DETECTION OF HLA GENES AND GENE PRODUCTS
A. MHC and HLA Classes
Six different loci of the HLA system have been identified and are divided in two classes (Table 3.1). Homologous MHC classes have been defined in other mammalian species.
MHC class I includes three major loci (HLA-A, HLA-B, HLA-C) and four minor loci (HLA-E, F, G, and H). Each locus has multiple serologically defined alleles, ranging from 8 in the case of HLA-C to more than 30 in the case of HLA-B. Each allele is designated by a number (e.g., HLA-B27). MHC class II antigens are equally polymorphic and include three main loci (HLA-DP, HLA-DQ, and HLA-DR) and less well-defined loci (HLA- DM, -DN, and -DO).
B. Structure of the MHC Antigens
1. Class I MHC Molecules
The HLA or H2 molecules coded by MHC class I genes have molecular weights which may vary from 43,000 to 48,000 and are formed by two nonidentical polypeptide chains. The major chain (α chain) has a long extracellular region folded in three domains—α1, α2, and α3. The extracellular domains are attached to a short transmembrane, hydrophobic region of 24 amino acids and an intracytoplasmic “tail” composed of about 30–35 amino acids, which includes the carboxyl terminus attached to cytoskeletal structures. β2-Microglobu-lin, a 12,000-dalton protein coded by a gene located on chromosome 15, is postsyntheti- cally and noncovalently associated with the major polypeptide chain.
The comparison of amino acid sequences and nucleotide sequences of the various do-mains of class I MHC shows that the α1 and α2 domains are highly variable and that most of the amino acid and nucleotide changes responsible for the differences between alleles occur in these domains. It also shows areas in these domains which are relatively constant and closely related in different alleles. This explains why polyclonal antibodies raised against MHC molecules can recognize several of them, an occurrence designated as the ex-istence of “public” specificities, as opposed to the “private” specificities unique to each dif-ferent allele.
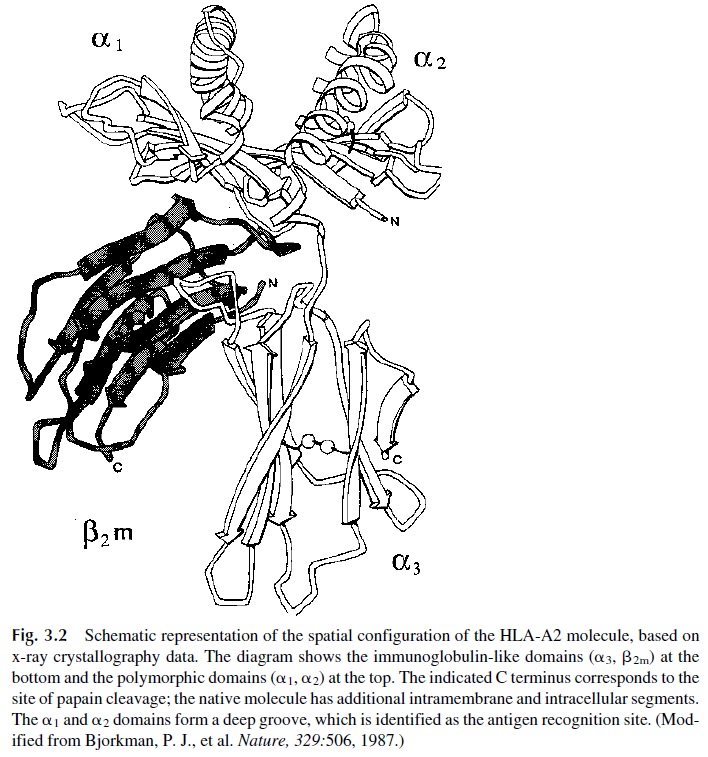
X-ray crystallography studies have determined the tridimensional structure of the HLA class I molecules (Fig. 3.2) and clarified the relationship between the structure and the function of this molecule. The most polymorphic areas of the molecule are located within and on the edges of a groove formed at the junction of the helical α1 and α2 domains. This groove is usually occupied by a short peptide (10–11 residues), usually of endogenous origin. The α3 domain shows much less genetic polymorphism and, together with β2-mi-croglobulin, are like a frame supporting the deployment in space of the more polymorphic α1 and α2 domains. In addition, the α3 domain has a binding site for the CD8 molecule characteristic of cytotoxic T cells.
2. Class II MHC Molecules
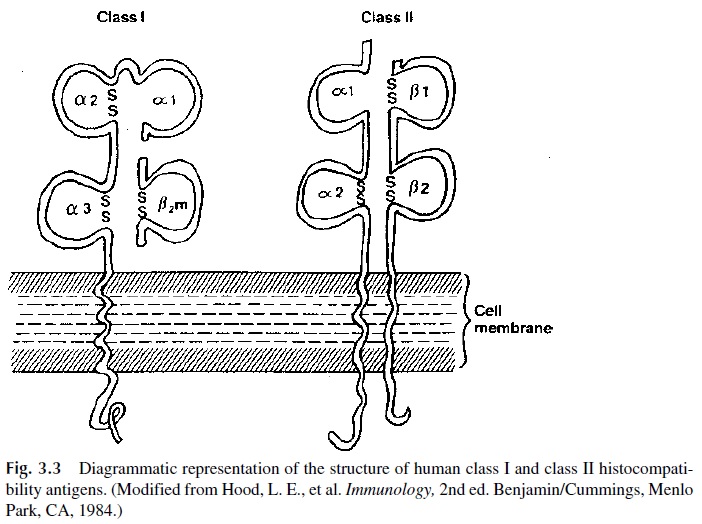
Although a remarkable degree of tertiary structure homology seems to exist between class I and class II gene products (Fig. 3.3), there are important differences in their primary struc-ture. First, class II gene products are not associated with β2-microglobulin. The MHC II molecules consist of two distinct polypeptide chains, a β chain (MW 28,000), which ex-presses the greatest degree of genetic polymorphism, and a less polymorphic, heavier chain (α chain, MW 33,000). Each polypeptide chain has two extracellular domains ( α1 and α2; β1 and β2), a short transmembrane domain, and an intracytoplasmic tail. The NH2 termi-nal ends of the terminal α1 and β1 domains contain hypervariable regions.
The three-dimensional structure of class II antigens has also been established. The β1 domains of class II MHC antigens resemble the α2 domain of their class I counterparts. The junction of α1 and β1domains forms a groove similar to the one formed by the α1 and α2 domains of class I MHC antigens, which also binds antigen-derived oligopeptides (usually of exogenous origin), but it is longer than the groove of MHC-I, indicating that the peptides bound to it are longer.
The β1 domain also contains two important sites located below the antigen-binding site. The first acts as a receptor for the CD4 molecule of helper T lymphocytes (see Chap-ters 4, 10, and 11). The second site, which overlaps the first, is a receptor for the envelope glycoprotein (gp 120) of the human immunodeficiency virus (HIV).
C. Identification of HLA Antigens
The different alleles of each locus are recognized by two major technique. The serological technique, which is the oldest and most widely used, is based on the lymphocytotoxicity of anti-HLA antibodies of known specificity in the presence of complement. The antibodies used for HLA typing were initially obtained from multiparous females or from recipients of multiple transfusions. Such antibodies are still in use, but monoclonal antibodies have also been raised against HLA specificities. These antibodies identify sev-eral groups of HLA that are therefore designated as serologically defined.
At this point it is important to note that not all HLA specificities have been defined. Some individuals express unknown specificities at some loci (usually class II), which the typing laboratory reports as “blank.” Investigation of these “blank” specificities often leads to the discovery of new HLA antigens. To avoid unnecessary confusion, they are assigned a numerical designation by regularly held workshops of the World Health Organization. At first, the designation is preceded by a w, indicating a provisional assignment. For example, DQw3 designates an antigenic specificity of the DQ locus that has been tentatively desig-nated as w3 by a workshop. When worldwide agreement is reached about the fact that this is a new specificity, the w is dropped.
Hybridization with sequence-specific oligonucleotides is particularly useful for typ-ing MHC-II specificities. A very large number of complete DNA sequences of class I and class II alleles have been determined. In the case of MHC-I molecules all alleles correspond to variations in the α chains, because β2-microglobulin is identical in all MHC-I molecules. In the case of HLA-DR molecules the α chain is invariant but the β-chain genes are ex-tremely polymorphic. In contrast, HLA-DP and DQ molecules have polymorphic α and β chains, thus being much more diverse than the DR molecules. As the sequences of HLA genes became known, it became possible to produce specific probes for different alleles. Typing usually involves DNA extraction, denaturation into single-stranded DNA, frag-mentation with restriction enzymes, amplification by polymerase chain reaction (PCR), and finally hybridization with labeled cDNA probes specific for different alleles of the cor-responding genes.
At the present time two partially overlapping sets of HLA alleles have been defined. One is serologically typed; the other is identified by nucleotide sequence. In the case of HLA-A, B, C, and DR, most nucleotide-defined alleles are subsets of the serologically de-fined ones. In the case of HLA-DP and DQ, half or more of the nucleotide-defined alleles do not have a known serological counterpart. The nomenclature of the nucleotide-defined alleles is somewhat confusing, although it obeys simple rules. For example, nucleotide-de-fined alleles of HLA-A1 are designated as HLA A*0101 and *0102, while nucleotide-de-fined alleles of HLA-B7 are designated as HLA-B*0702-*0706. In the case of DQ and DP the nomenclature identifies both the polypeptide chain where the nucleotide sequence has been identified and the serological corresponding marker, if defined. For example, the HLA-DQB1*0501 allele is a nucleotide-defined allele associated with the β chain of the serologically defined DQ5 allele.
Related Topics