Chapter: Biotechnology Applying the Genetic Revolution: DNA, RNA, and Protein
Translating the Genetic Code into Proteins
TRANSLATING THE GENETIC CODE INTO PROTEINS
The Genetic Code Is Read as Triplets or Codons
Messenger RNA provides the information a ribosome needs to make proteins. This process is known as translation because it involves translating information carried by nucleic acids to give the sequences of amino acids that make up proteins. Before the mechanism is discussed, the code used to assemble proteins must first be understood. Nucleic acids each have four different bases (the T in DNA is equivalent to U in RNA). However, proteins consist of 20 different amino acids. If each nucleotide corresponded to an amino acid, this would encode only four different amino acids. Two nucleotides give only 16 combinations, still not enough. To encode 20 amino acids, bases must be read in groups of three. This gives 64 different combinations— more than enough for all 20 amino acids (Fig. 2.15). Messenger RNA is therefore read in groups of three bases, known as triplets orcodons. Each triplet of bases codes for one amino acid. Because there are more than 20 triplets, many are redundant so that multiple codons will be translated into the same amino acid. For example, valine is encoded by GUU, GUC, GUA, or GUG.
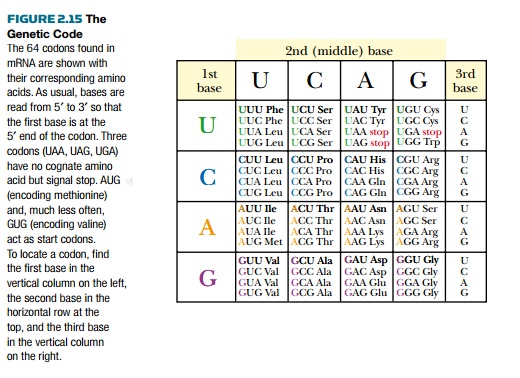
The genetic code listed in Fig. 2.15 is considered the universal genetic code. Not all organisms use precisely this code, although exceptions are rare. For example, UGA normally signals stop, but in Mycoplasma, UGA encodes tryptophan and, in the protozoan Euplotes, UGA encodes cysteine.
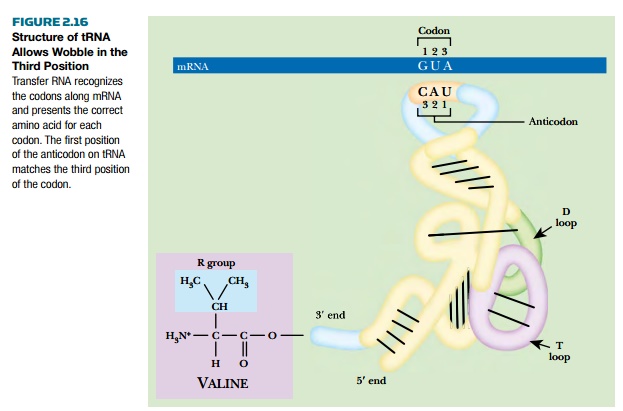
Small RNA molecules known as transfer RNA (tRNA) recognize the individual codons on mRNA and carry the corresponding amino acids. Although tRNA is synthesized as a single strand, it folds back on itself to form regions of double-stranded RNA. The final shape of tRNA is a folded “L” shape with the anticodon at one end and the acceptor stem at the other. The anticodon consists of three bases complementary to those of the corresponding codon and it therefore recognizes the codon by base pairing. The acceptor stem is where the amino acid is added to the free 3′ end of the tRNA (Fig. 2.16).
How does each specific tRNA carry the correct amino acid?
A group of enzymes called aminoacyl tRNA synthetases attach the correct amino acid to the corresponding tRNA. These enzymes are very specific and recognize the correct tRNA by its sequence at the anticodon or elsewhere along the RNA structure. There is a specific aminoacyl tRNA synthetase for each amino acid.
The first base of the anticodon binds the third base of the codon in the mRNA. Because this nucleotide in tRNA is not constrained by neighboring nucleotides, it can wobble instead of forming a perfect double helix. This allows nonstandard base pairs to be created. For example, if the first anticodon base is G, it would normally pair with C in the third position of the codon. Because of wobble, G can also pair with U. Thus, the tRNA for histidine has the anticodon GUG and recognizes both CAC and CAU in the mRNA. Similarly, U in the first place in the anticodon can base-pair with A or G in the third position of the codon. Wobble explains how the same tRNA can read multiple codons all encoding the same amino acid.
Protein Synthesis Occurs at the Ribosome
The ribosome is a molecular machine that unites the mRNA with the appropriate tRNAs and then links the amino acids together into a chain. Prokaryotic ribosomes consist of two subunits called the 30S and 50S, which combine to form a functional 70S ribosome. A ribosome consists of several RNA molecules (ribosomal RNA or rRNA) and many proteins. The 30S subunit has a 16S rRNA plus 21 proteins; the 50S subunit has two rRNAs, the 5S and 23S, plus 34 proteins. The larger subunit has three binding sites for tRNA, called A for acceptor, P for peptide, and E for exit, referring to the action occurring at each site. The 23S rRNA actually catalyzes the addition of amino acids to the growing polypeptide chain and is therefore a ribozyme.
In prokaryotes, various factors beside the ribosome are involved in protein synthesis (Fig. 2.17). First, a ribosome must assemble at the start site and begin protein synthesis at the correct start codon. The 5′ untranslated region of the mRNA (see above) has the signal for ribosome binding in front of the start codon. In prokaryotes, translation begins at the first AUG codon after the Shine-Dalgarno sequence, or ribosome binding site, which has the consensus sequence UAAGGAGG. The anti-Shine-Dalgarno sequence is found in the 16S rRNA of the smaller 30S subunit. So first, the small ribosomal subunit binds the Shine-Dalgarno sequence. A derivative of methionine, N-formyl-methionine (fMet), and a special initiator tRNA(tRNAi) are used to initiate translation in prokaryotes. Only initiator tRNA charged with fMet (referred to as tRNAifMet) can bind the small subunit of the ribosome.
Translation factors are proteins needed to recruit and assemble the components of the ribosome and translational complex. Initiation factors (IF1, IF2, and IF3) assemble the 30S initiation complex, which is the 30S ribosomal subunit plus tRNAifMet. The IF3 factor then leaves the complex, and the 50S ribosomal subunit binds, forming the 70S initiation complex (see Fig. 2.17A).
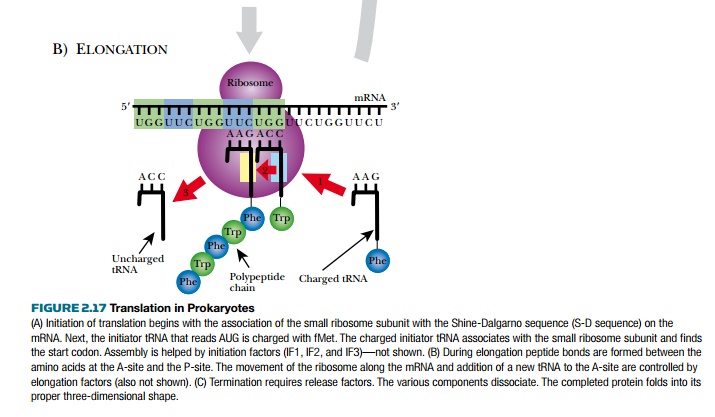
Finally, polypeptide assembly can begin (see Fig. 2.17B). The tRNA fmet occupies the P- site on the ribosome. Another tRNA recognizes the next codon and enters the A-site, and a peptide bond is formed between the first and second amino acid by the peptidyl transferase activity of 23S rRNA. fMet releases its tRNA, which moves into the E-site. This allows the second tRNA to move into the P-site, and the cycle begins again. A third tRNA, complementary to the next codon, enters the A-site, a peptide bond forms between amino acids 2 and 3, and then the second tRNA moves into the E-site of the ribosome, and exits.
Adding successive amino acids is called elongation and requires elongation factors. EF-T is a pair of proteins (EF-Tu and EF-Ts) that uses GTP to catalyze the addition of a new tRNA into the A-site (EF-Tu) and then exchanges the GDP with GTP for the next cycle (EF-Ts).
The movement of tRNA from the P-site to the E-site is called translocation, and the mRNA simultaneously moves one codon sideways relative to the ribosome. The E-site and A-site cannot be occupied at the same time, and the used tRNA must exit before the next tRNA enters. EF-G oversees the translocation step.
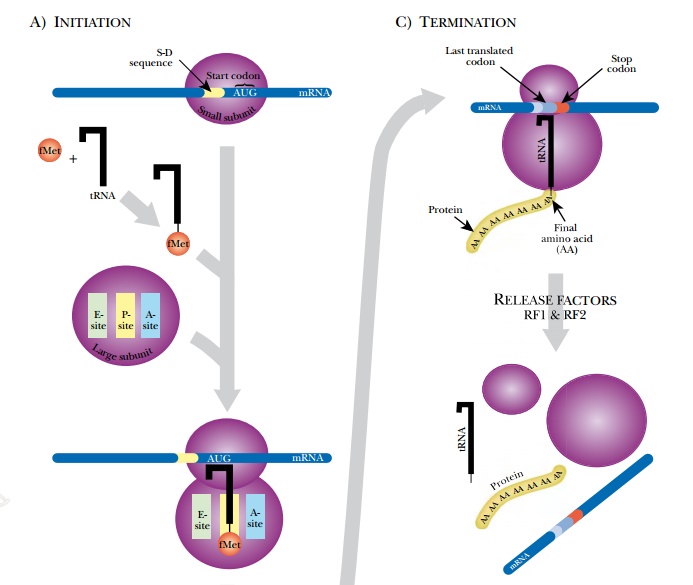
Amino acids are added to the growing chain and the process continues until the ribosome encounters a stop codon (UAA, UAG, or UAA). None of the tRNA recognizes the stop codon. Instead proteins known as release factors bind the stop codons (see Fig. 2.17C). RF1 and RF2 recognize the different stop codons and stimulate the 23S rRNA to split the bond between the last amino acid and its tRNA. The whole ribosome assembly falls off the mRNA and dissociates. Its components will be used again for translation of another mRNA. The new polypeptide chain folds to form its final structure. In prokaryotes, multiple ribosomes bind to the same mRNA to form a polysome. Because there is no nucleus, transcription and translation are often simultaneous.
As partially made mRNA comes off the DNA, ribosomes bind and start synthesizing protein.
Related Topics