Chapter: Biotechnology Applying the Genetic Revolution: DNA, RNA, and Protein
Regulation of Transcription in Eukaryotes
REGULATION OF TRANSCRIPTION IN EUKARYOTES
Just as the initiation of transcription is more complex in eukaryotes, so is its control. The mechanisms to regulate which gene is expressed at what time are very complicated. The fact that eukaryotic DNA is wound around histones hinders many proteins from binding to the DNA, delaying access by activators and repressors. In addition, the nuclear membrane prevents the access
of most proteins to the nucleus. Consider the complexity of the human body, with its multiple tissues. Each gene in each cell needs to be expressed only when needed, and only in the amount needed. In addition to normal organ functions and to changes during development, the environment has a huge impact on our bodies, and changes in gene expression help us adapt. Overall, the numbers and types of controls for gene expression are staggering. Other eukaryotes such as mice, rats, Arabidopsis, C. elegans, and even the relatively simple yeast have similarly complex control systems.
There are many different transcription factors for eukaryotic genes, yet they all have at least two domains: one binds to DNA, and the other binds to some part of the transcription apparatus. The two domains are connected, yet may function when separated from each other (Fig. 2.10).
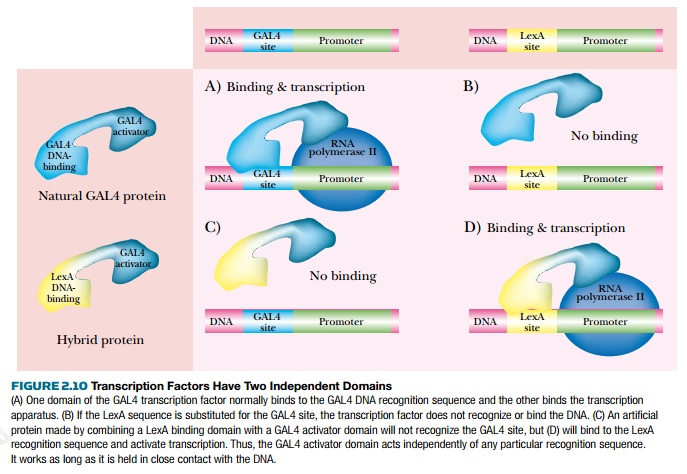
If the DNA binding domain of one transcriptional regulator is connected to the activation domain of another, the hybrid protein will work, each part retaining its original characteristics. That is, the DNA binding domain will bind the same sequence as it did before, and the activation domain will activate transcription as before. This property can be exploited when trying to identify protein-to-protein interactions with newly characterized proteins in the yeast two-hybrid screen.
Transcription factors work via an assembly of many proteins called the mediator complex (Fig. 2.11). These proteins receive all the signals from each of the activator proteins, compile the message, and transmit this to RNA polymerase II. The mediator contains about 20 different proteins, most of which make up the core. The presence of other proteins
may vary depending on the cell or organism. These accessory proteins were originally thought to be co-activators or co-repressors since their presence varies based on tissue.
The mediator complex sits directly on RNA polymerase II waiting for information from activators or repressors. These may bind to regions just upstream of RNA polymerase II and the mediator complex. However, eukaryotic transcription factors may also bind to DNA sequences known asenhancers that may be thousands of base pairs away from the promoter. Even so, the regulatory proteins bind directly to the mediator complex. The enhancer elements work in either orientation, but affect only genes that are in the general vicinity. The prevailing theory is that the DNA loops around so that the enhancer is brought near the promoter.
Insulators are DNA sequences that prevent enhancers from activating the wrong genes. Insulators are placed between enhancers and those genes they must not regulate. The insulator binding protein (IBP) recognizes the insulator sequences and blocks the action of enhancers that are on the far side of the insulator (see Fig. 2.11). Insulator sequences may be controlled by methylation. When the DNA sequence is methylated, IBP cannot bind and the enhancer is allowed to access promoters beyond the insulator.
The eukaryotic transcription factor has two domains. The DNA binding domain binds to the DNA at the pro-moter and the activator domain has sites for initiating RNA polymerase action.
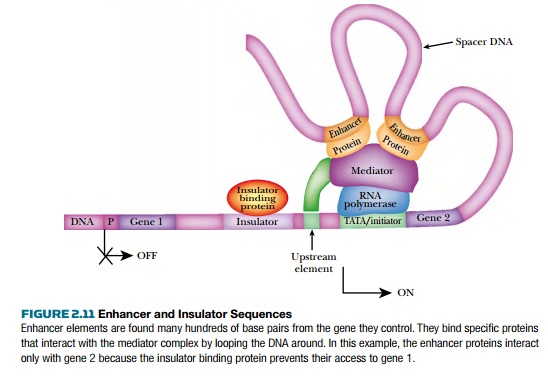
Eukaryotic Transcription Enhancer Proteins
An example of a eukaryotic transcription factor that works as an activator in some cell types is AP-1 (activator protein-1). This affects a variety of genes and responds to a wide range of different stimuli. The most potent stimulators of AP-1 include growth factors and UV irradiation. Growth factors stimulate cell growth, and UV irradiation kills cells, so the genes activated by AP-1 are quite varied. The complex effects of this single transcription factor are still being investigated.
AP-1 is a dimer of two proteins, Fos and Jun, and recognizes the palindromic sequence 5′-TGACTCA-3′ (Fig. 2.12). AP-1 belongs to a family of DNA binding proteins called bZIP proteins. The proteins each have a leucine zipper, which brings the partners
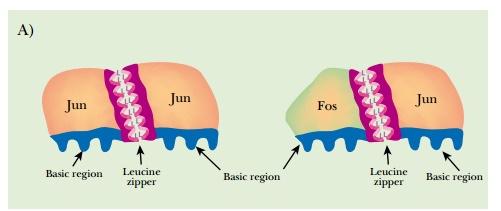
together to form dimers, plus a region of basic residues that contacts the DNA. (Leucine zippers are alpha helical segments where one side of the helix is mostly leucine. Two such leucine patches will associate by hydrophobic bonding and can bind two different proteins together.) Jun can either dimerize with itself or bind to Fos. In contrast, Fos can bind to Jun but cannot dimerize by itself. Fos and Jun also have activation domains that receive cellular signals that increase or attenuate their activity. Other protein partners may bind to Fos and Jun, and these change the recognition sequence. These other proteins, such as ATF, Maf, and Nr1, add to the complexity of AP-1 transcription control.
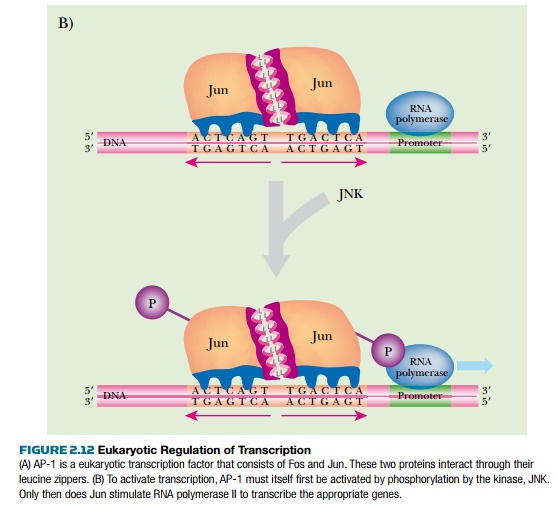
When AP-1 is stimulated, two different effects are involved. First, the cell makes more Fos and Jun proteins through increased expression of their genes. In addition, the proteins themselves become more stable and are not degraded as quickly. Second, the activity of Fos and Jun are stimulated by phosphorylation of their activation domain by JNK (Jun amino-terminal kinase). Many other cellular signaling proteins can alter Jun and Fos activity, but JNK is the most potent. Phosphorylation of Jun and Fos triggers their interaction with the protein mediator complex and RNA polymerase II. It also affects other signal proteins and triggers other genes.
The eukaryotic transcription factor, AP-1, consists of two different proteins that work as a dimer. The family of proteins that form AP-1 are also controlled by posttranslational modifications such as phosphorylation.
DNA Structure Affects Access of Proteins to Promoter Regions
As noted earlier, the structure of eukaryotic DNA has a huge impact on the ability of regulatory proteins to bind. During interphase, eukaryotic chromosomes are uncondensed and looped, with the ends of the loops attached to the nuclear matrix.
This helps the binding of transcription factors. The amount of condensation into heterochromatin is a critical factor. Eukaryotic DNA is wrapped around histones, forming nucleosomes.
Loosely packed nucleosomes provide access for transcription factors, whereas tightly packed nucleosomes exclude all other proteins. Therefore, controlling the density of nucleosomes can regulate transcription initiation.
The histone proteins have tails that can be acetylated by enzymes called histone acetyl transferases (HATs). These histone tails normally stabilize DNA by binding neighboring nucleosomes, so aggregating the nucleosomes. When HAT transfers acetyl groups to the tails, they can no longer bind to neighboring nucleosomes, and the structure loosens. In order to tighten the nucleosomes, histone deacetylases (HDACs) remove the acetyl groups, and the histones reaggregate (Fig. 2.13).
Another structural feature of eukaryotic DNA important to gene expression is methylation (see Fig. 2.13). In prokaryotes, differences in methylation patterns distinguish the newly synthesized strand of DNA from the template during replication. In eukaryotes, methylation is used to silence various regions of DNA and prevent their expression. Methylation of cytosine by two different enzymes occurs in the sequence CG or CNG. Maintenance methylases add methyl groups to newly synthesized DNA to give the same pattern as in the template strand. De novo methylases add new methyl groups, and of course, demethylases remove unwanted methyl groups. Many genes are located near stretches of DNA containing many CG sequences, called CG islands. If these are methylated, the nearby genes are not expressed, whereas if they are not methylated, the genes are expressed. Methylation patterns depend on the tissue. For example, muscle cells will not methylate CG islands in front of genes necessary for muscle function. The muscle-specific genes will have methylated CG islands in other tissues, though.
Silencing by DNA methylation can occur for one gene or for areas as large as an entire chromosome. Regions that become “silenced” first have methylated CG regions.
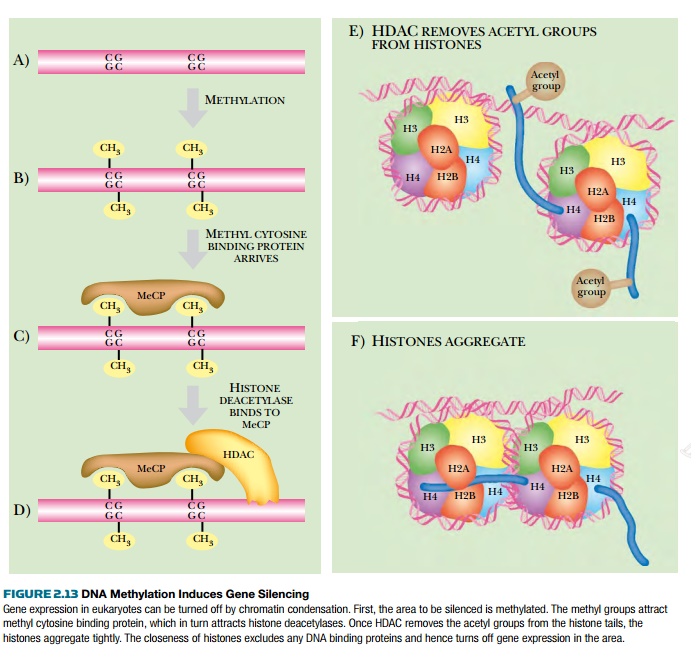
The methylated DNA attracts methylcytosine binding proteins that block binding of other DNA binding proteins. These proteins also recruit HDACs that deacetylate the histone tails, thus condensing the chromatin. The areas become heterochromatin, and this prevents any further gene expression.
Methylation patterns have a major impact during development, because gene expression here must be tightly controlled. Some genes remain methylated in the gamete, whereas other genes must be demethylated so they can become active. Imprinting occurs when a gene from the parent is methylated in the gamete and remains methylated in the new organism. This affects relatively few genes. In contrast, genes that are not imprinted change their methylation patterns during development. Imprinting can be different for male gametes
and female gametes, setting the stage for sexual differences in gene expression. One special type of imprinting is X-inactivation, where the entire second X chromosome in females is silenced by methylation (except for a few loci).
Related Topics