Chapter: Plant Biochemistry: ATP is generated by photosynthesis
The synthesis of ATP is effected by a conformation change of the protein
The synthesis of ATP is effected by a conformation change of the protein
For the reaction:
ATP + H 2O ← → ADP + Phosphate
the standard free energy is:
ΔG°’ = -30.5 kJ/mol
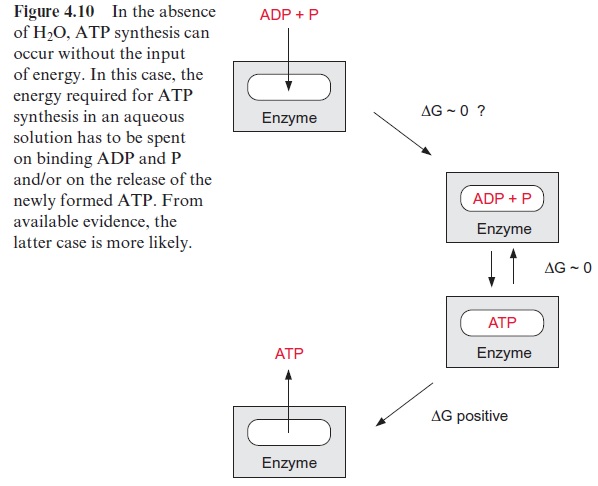
Because of its high free energy of hydrolysis, ATP is regarded as an energy-rich compound. It should be noted, however, that the standard value ΔG°’ has been determined for an aqueous solution of 1 mol of ATP, ADP, and phosphate per liter, respectively, corresponding to a water con-centration of 55 mol/L. If the concentration of water was only 10 -4 mol/L, the ΔG for ATP hydrolysis would be +2.2 kJ/mol. This means that at very low concentrations of water the reaction proceeds towards the synthesis of ATP. This example demonstrates that in the absence of water the synthesis of ATP does not require the uptake of energy.
The catalytic site of an enzyme can form a reaction site where water is excluded. Catalytic sites are often located in a hydrophobic area of the enzyme protein in which the substrates are bound in the absence of water. Thus, with ADP and P tightly bound to the enzyme, the synthesis of ATP could proceed spontaneously without requiring energy (Fig. 4.10). This has been proved for H+ -ATP synthase. Since the step of ATP synthesis itself does proceed without the uptake of energy, the amount of energy required to form ATP from ADP and phosphate in the aqueous phase has to be other-wise consumed, e.g., for the removal of the tightly bound newly synthesized ATP from the binding site. This could occur by an energy-dependent con-formation change of the protein.
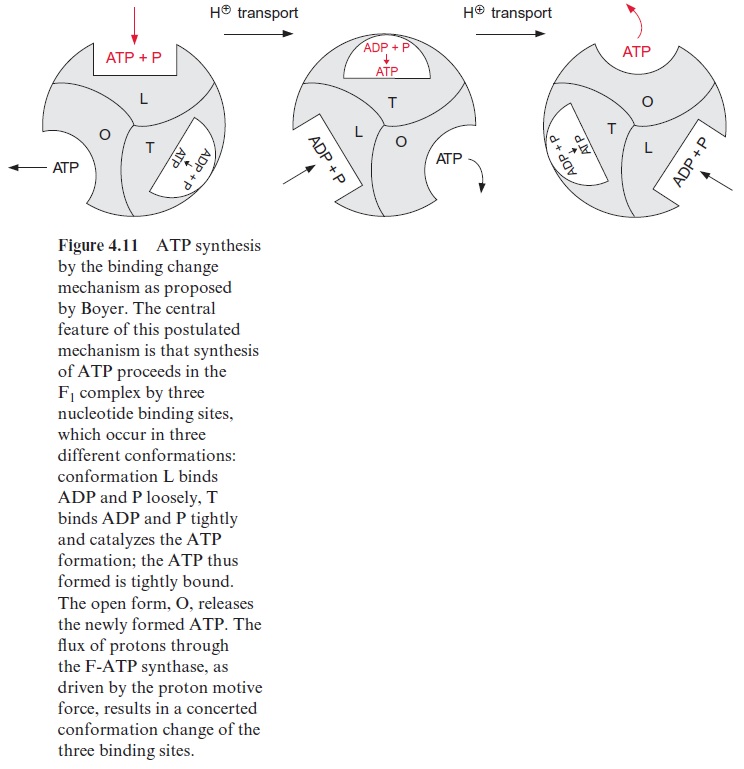
In 1977 Paul Boyer (USA) put forward the hypothesis that the three identical sites of the F1 protein alternate in their binding properties (Fig. 4.11). One of the binding sites is present in the L form, which binds ADP and phosphate loosely but is not catalytically active. A second binding site, T, binds ADP and ATP tightly and is catalytically active. The third binding site, O, is open, binds ADP and ATP only very loosely, and is catalytically inactive. According to this “binding change” hypothesis, the synthesis of ATP proceeds in a cycle. First, ADP and phosphate are bound to the loose binding site, L. A conformational change of the F1 protein converts site L to a binding site T, where ATP is synthesized from ADP and phosphate in the absence of water. The ATP formed remains tightly bound. Another conformational change converts the binding site T to an open binding site O, and the newly formed ATP is released. A crucial point of this hypoth-esis is that with the conformational change of the F1 protein, driven by the energy of the proton gradient, the conformation of each of the three cata-lytic sites is converted simultaneously to the next conformation
(L → T T → O O → L)
The results of X-ray analysis, shown above, support the binding change hypothesis. The evaluated structure clearly shows that the three subunits of F1—one free, one loaded with ADP, and one with the ATP analogue AMP-PNP—have different conformations. Paul Boyer and John Walker were awarded the Nobel Prize (1997) for these results. However, the details of this mechanism are still a matter of debate.
Further investigations showed that the central γ-subunit rotates. The γ-and ε-subunits of F1 together with the 12 c-subunits of Fo (shown in red in Fig. 4.7) form a rotor. This rotor rotates in a stator consisting of subunits-(αβ)3, , a, b, b’ , by which the conformations of each of the catalytic cent-ers shown in Fig. 4.11 is changed. This model suggests that three molecules of ATP are formed by one complete revolution.
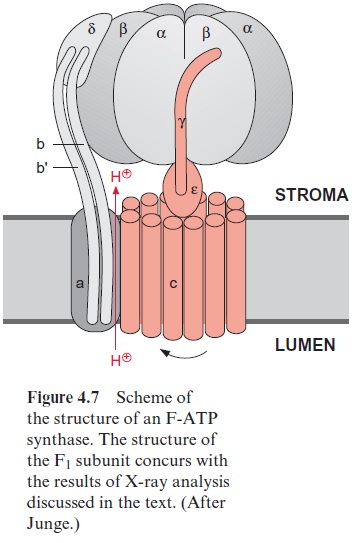
This model was confirmed by a startling experiment carried out by Masasuka Yoshida and Kazohiko Kinosito in Japan. These researchers attached a fluorescent molecule to the upper end of a γ-subunit contained in an Fo particle present in a membrane. Using a special video microscopy documentation it was possible to make visible that upon the hydrolysis of ATP the γ-subunit did in fact rotate. The Fo part functions as a type of nano motor. The velocity of rotation of the F-ATP-synthase in chloroplasts has been estimated to be up to 160 revolutions per second. To explain how this nano motor is driven by a proton gradient on the basis of known struc-tural data, Wolfgang Junge (Osnabrueck, Germany) developed the model shown in Figure 4.12. In this model the a-subunit of the stator (shown in gray) possesses a channel through which protons can move from the out-side of the membrane to a binding site of a c-subunit of the rotor, possibly a glutamate residue. At another site of the stator is a second channel through which the protons bound to the c-subunit can be released to the inside. By Brownian movement this proton-loaded c-subunit can rotate to the other proton channel where the proton is released, facilitating a proton transport driven by the proton gradient from the outside to the inside. But why does the rotation caused by Brownian movement proceed only in one direc-tion? Two factors could be responsible for this: the spacial separation of the two channels and a positively charged arginine residue of the a-subunit of the stator. It is assumed that the positive charge of the arginine repels the proton-loaded subunit and thus prevents a backward movement of the rotor, orientating the Brownian movement into one direction. Driven by a proton gradient that causes the loading and unloading of the proton bind-ing sites at the respective channels, according to this model the nano motor rotates step by step like a ratchet in only one direction. In this way one full revolution causes the conformational change in the F1-part leading to the formation of three molecules of ATP. Although an experimental veri-fication of this model remains to be done, it gives an idea of how the nano motor of the ATP synthase may be driven by a proton gradient.
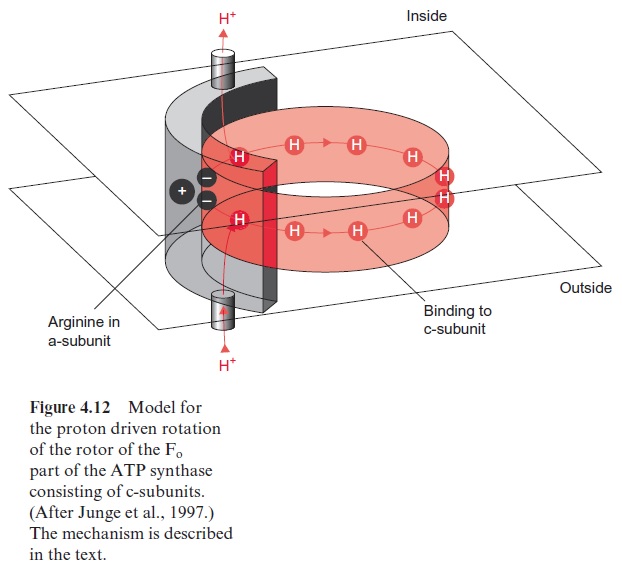
As discussed previously, several bacteria contain an F-ATP synthase that is driven by an Na+ gradient. The model of the proton driven rotor allows the assumption that the subunit c of the Na+ F-ATP synthase binds Na+ ions and the two partial ion channels conduct Na+.
It is still unclear how many c-subunits make up the rotor. Investigations of the number of c-subunits per F-ATP synthase molecule yielded values of 12 to 14 (chloroplasts), 10 (yeast mitochondria), and 12 (E. coli). Apparently in various organisms the number of c-subunits in the Fo part vary, therefore the number of protons required for one revolution to form three molecules of ATP will vary accordingly.
In photosynthetic electron transport the stoichiometry between the formation of NADPH and ATP is still a matter of debate
According to the model discussed here, chloroplasts with 14 c-subunits per rotor would require 14 protons for a complete rotation. Since three ATP mol-ecules are formed during one rotation, this would correspond to an H+ /ATP ratio of 4.7. Independent measurements indicated that in chloroplasts at least four protons are necessary for the synthesis of one ATP, which would be sim-ilar to the proton stoichiometry calculated for the rotor model. It is still not clear to what extent the Q-cycle plays a role in proton transport. In the linear (noncyclic) electron transport, for each NADPH formed without a Q-cycle, four protons (PS II: 2H+ , Cyt-b6/f complex: 2H+ ), and with a Q-cycle (Cyt-b6/f complex: 4H+ ) six protons, are transported into the lumen . With a H+ /ATP ratio of 4.7, for each NADPH produced 1.3 ATP would be generated with the Q-cycle in operation and just 0.9 ATP without a Q-cycle. If these stoichiometries are correct, noncyclic photophosphorylation would not be sufficient to meet the demands of CO2 assimilation in the Calvin cycle (ATP/NADPH=1.5) and therefore cyclic photophosphoryla-tion would be required as well. The question concerning the stoi-chiometry of photophosphorylation is still not finally answered.
H -ATP synthase of chloroplasts is regulated by light
H+ -ATP synthase catalyzes a reaction that is in principle reversible. In chlo-roplasts, a pH gradient across the thylakoid membrane is generated only during illumination. In darkness, therefore, due to the reversibility of ATP synthesis, one would expect that the ATP synthase then operates in the opposite direction by transporting protons into the thylakoid lumen at the expense of ATP. In order to avoid such a costly reversion, chloroplast ATP synthase is subject to strict regulation. This is achieved in two ways. If the pH gradient across the thylakoid membrane decreases below a threshold value, the catalytic sites of the β-subunits are instantaneously switched off, and they are switched on again when the pH gradient is restored upon illumination. The mechanism of this is not yet understood. Furthermore, chloroplast ATP synthase is regulated bythiol modulation. By this process, a disulfide bond in the γ-subunit of F1 is reduced in the light by ferredoxin to form two -SH groups. This is mediated by reducedthioredoxin. The reduction of the γ-subunit causes the activation of the catalytic centers in the β-subunits. In this way illumination switches F-ATP synthase on. Upon darkening, the two -SH groups are oxidized by oxygen from air to form a disulfide, and as a result of this, the catalytic centers in the β-subunits are switched off. The simultaneous action of the two regulatory mechanisms allows an efficient control of ATP synthase in chloroplasts.
V-ATPase is related to the F-ATP synthase
Vacuoles contain a proton which transports V-ATPase and is conserved in all eukaryotes. Some V-ATPases transport Na+ ions instead of protons. In plants, V-ATPases are located not only in vacuoles (V vacuoles), but also in plasma membranes and membranes of the endoplasmic reticulum and the Golgi apparatus. Genes for at least 12 different subunits have been iden-tified in Arabidopsis thaliana. Major functions of this pump are to acidify the vacuole and to generate proton gradients across membranes for driv-ing the transport of ions. V-ATPases also play a role in stomatal closure of guard cells. They resemble the F-ATP synthase in its basic structure, but are more complex. They consist of several proteins embedded in the mem-brane, similar to the Fo part of the F-ATPase, to which a spherical part (e.g., F1) is attached by a stalk that protrudes into the cytosol. The spheri-cal part consists of 3A- and 3B-subunits, which are arranged alternately like the (αβ)-subunits of F-ATP synthase. F-ATP synthase and V-ATPase are derived from a common ancestor. The exact number of protons transported per ATP depends on how many c-subunits the rotor of the Fo part contains. The V-ATPase is able to generate titratable proton concentrations of up to 1.4 mol/L within the vacuoles.
Vacuolar membranes also contain an H+ -pyrophosphatase, which upon the hydrolysis of one molecule of pyrophosphate to phosphate pumps one proton into the vacuole, but it does not reach such high proton gradients as the V-ATPase. H+ -pyrophosphatase probably consists of only a single pro-tein with 16 transmembrane helices. It remains to be elucidated why there are two enzymes transporting H+ across the vacuolar membrane. Plasma membranes contain a proton transporting P-ATPase.
Related Topics