Chapter: Biotechnology: Microbial Cell Culture and its Applications
Strain isolation, improvement and preservation
Strain isolation, improvement and preservation
By this time, you must have realized that one of our major purposes to culture microbes is to produce useful products. But, do you think that all microbes produce useful or novel products? The answer is no. So, we have to search for those microbes, which produce substances of our interest. Where do we get these microorganisms?
As you know, microbes are wide spread in natural habitats especially in soil and in aquatic environments. They are also found in extreme environments namely arctic waters and hot springs. These natural habitats are our source of microbes. Once we have isolated microbes of desired interest, we can further improve their desired traits using a variety of methods.
Strain isolation:
The sample containing the microbes (e.g. soil) is put in a nutritive medium and allowed to grow in shake cultures. The growth conditions (e.g. temperature) or nutrients in the medium are provided such that these favour the growth of microbes of our interest. This is called enrichmenttechnique. The enriched culture can further be sub-cultured by taking a small inoculum andputting it into fresh medium. In this way, the growth of the desired organisms improves successively. Further screening is done using a method where the organism will show its desired properties. For example, if we are looking for a microorganism, which produces an antibiotic, we may detect it by growing the culture on an agar plate in the presence of that bacterium against which antimicrobial activity is desired. Immunological methods are also available in which the microbes producing products are detected using specific antibodies. Molecular biology has made available a variety of probes, which enable the detection of organisms capable of producing specific products. Recently some of these methods have been adapted to robotic automation resulting in enormous throughput screening of microbes for newer / novel molecules.
Strain improvement:
Strain isolation procedure described above only identifies a strain, which has the capability or potential to produce a desired molecule. It does not ensure that it produces molecule in sufficient quantities to be economically viable. Techniques of classical genetics and genetic engineering are used to improve the desirable characteristics of the strain.
Mutation Selection: This is one of the oldest methods of strain improvement. The strain isexposed to chemical (e.g. nitrosoguanidine or NTG) or physical (e.g. UV rays) mutagens and the mutants having improved characteristics are selected. It is often necessary to carry out multiple successive mutations before we get the desired results. One of the classical examples of strain improvement using this methodology is the production of antibiotic penicillin. Several successive mutations were necessary to develop a strain of Penicilliumchrysogenum capable of producing nearly 100 times the concentration of penicillin produced by the original strain (Penicilliumnotatum), thus making production of penicillin commercially feasible.
Genetic Engineering Techniques: Until the recent breakthroughs in the techniques of geneticengineering, a bacterium could produce only substances coded for in its genome. Genetic engineering techniques about which you have learnt allow totally new properties or capabilities to be added to the microorganisms giving rise torecombinant strains. Using these techniques, microorganisms may be manipulated to, synthesize or secrete enhanced quantities of biomolecules, facilitate production of novel compounds or allow utilization of cheaper substrates. Using these techniques, the microorganisms may also be utilized to produce plant, animal or human proteins. Some of the valuable human proteins which are being produced in microorganisms using this technology include recombinant human insulin (Humulin), hepatitis B surface antigen, human growth hormone and interferons. These proteins can now be produced in large quantities. Consequently the cost of the therapies which make use of these proteins viz. insulin (diabetes), hepatitis B surface antigen (vaccination against hepatitis B virus), human growth hormone (growth retardation) and interferons (immunotherapy) has been reduced considerably.
The tools, which are used for genetic engineering viz. the restriction enzymes, cloning and expression vectors and introduction of recombinant DNA into host cells. However there are many practical problems, which must be taken care of before a foreign (heterologous) gene may be expressed in a microorganism to make it commercially viable. For example, when a foreign gene is introduced into a host bacterium, it may not be expressed there. This problem is overcome by placing foreign gene under regulatory controls recognized by the host microorganism. To maximize production of foreign protein, the expression vector used is such that it replicates tohigh copy number and is stable. The foreign gene should ideally be linked to a strong promoter that has high affinity for RNA polymerase. The foreign gene may also be put under the control of a regulatory switch such that production of recombinant protein does not occur until required.
When a eukaryotic gene (e.g., plant, animal, human) is expressed in prokaryotic (bacterial) host, there are additional problems to be tackled. The non-coding region of eukaryotic gene must be excised. This requires use of reverse transcription of mRNA into cDNA. Additionally, therecombinant protein may not be secreted into the medium or its incorrect folding andaccumulation intracellularly may generate inclusion bodies. All these problems makedownstream processing difficult and costly. Thus, an alternative would be to use a eukaryotic expression host. For this purpose, Saccharomyces cerevisiae has been quite popular because it is safe and scientists have long experience of using this yeast in industrial fermentations. Detailed information on biochemistry, physiology and genetics of this yeast is also known. Moreover, this yeast can be manipulated genetically rather easily. However, product yields are relatively low at 1-5% of the total protein. Other yeasts like Pichiapastoris has a number of advantages: it has strong inducible promoters; it is capable of making post-translational modifications similar to those performed by human cells; downstream processing is simpler as Pichia does not secrete its own proteins into the fermentation medium.
Metagenomics
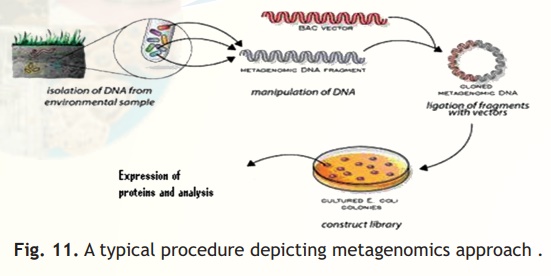
In the last few years, another approach has been developed to identify and select microbial genes synthesizing novel molecules. This approach directly utilizes the large number of microbial genomes present in an environmental niche, for example in soil, in water such as ocean or in human gut. These genomes are contributed by both the culturable and the non-culturable variety of microbes and together constitute what has been termed as metagenome. The collective DNA is extracted from a sample of soil, water or any other environmental niche. It is subjected to restriction digestion using restriction endonucleses and the fragments are cloned. The clones are then screened for presence of a variety of molecules. The clones expressing novel molecules or molecules with improved characteristics are used for large-scale production by fermentation techniques.
The metagenomic approach not only give the scientists an opportunity to cast a wider net on microbial resource present in the environment to fish out genes of their interest, it also gives them the opportunity to analyze the genomes of the microbes without culturing these in the laboratory. Thus, it is really a very useful approach to study those microbes, which are difficult to culture in the laboratory or have never been cultured in the laboratory as yet, and analyze these to see if they carry any genes, which may be exploited for human use. A typical procedure depicting metagenomic approach is shown in Fig. 11.
Strain preservation:
Once a strain producing a novel or desired product has been obtained, it must be appropriately preserved for future use. If not done properly, the strain may be lost through loss of viability or even show decline in the production of the product for which it was isolated.
Storage on agar: Cultures are grown on agar slants or stabs & stored at 5 to -20oC. These must besub-cultured at approximately 6-month interval. The time of sub-culture may be extended to 1 year if cultures are covered with sterile mineral oil.
Storage in liquid nitrogen: The culture is grown and a cryoprotective agent like glycerol (10-30%)is added. These are dispensed in sealed ampoules & frozen in liquid nitrogen. (-176 to -196 oC).
Lyophilisation: Lyophilization or freeze-drying involves freezing of a culture followed by dryingunder vacuum. This results in sublimation of cell water. Lyophilised culture may remain viable for 5-10 years or more.
Culture Collections Centers
Cultures may be deposited to culture collection centers. These centers safely maintain cultures for years. The cultures are also made available to prospective investigators. With the advent of the modern biotechnology and the associated commercial and financial implications, the culture collection centers are governed by stringent rules & regulations to protect the intellectual property rights of the depositors. Some of the well-known culture collection centers of international repute are ATCC (American Type Culture Collection, USA), NCIB (National Collection of Industrial Bacteria, UK) and DSM (Deutsche Sammlung von Mikroorganismen and Zelkulturen, Germany). The National culture collection of India is called MTCC (Microbial Type Culture aCollection and Gene Bank) and is located at Institute of Microbial Technology, Chandigarh.Recently another National culture collection centre named NBAIM (National Bureau ofAgriculturally Important Microorganisms) has been established in India at Mau in Uttar Pradesh(U.P.)
Related Topics