Chapter: Biochemistry: Protein Synthesis: Translation of the Genetic Message
Prokaryotic Translation
Prokaryotic Translation
The
details of the chain of events in translation differ somewhat in prokaryotes
and eukaryotes. Like DNA and RNA synthesis, this process has been more
thoroughly studied in prokaryotes. We shall use Escherichia coli as our principal example, because all aspects of
protein synthesis have been most extensively studied in this bacterium. As was
the case with replication and transcription, translation can be divided into
stages-chain initiation, chain elongation, and chain termination.
Ribosomal Architecture
Protein
synthesis requires the specific binding of mRNA and aminoacyl-tRNAs to the
ribosome. Ribosomes have a specific architecture that facilitates the binding.
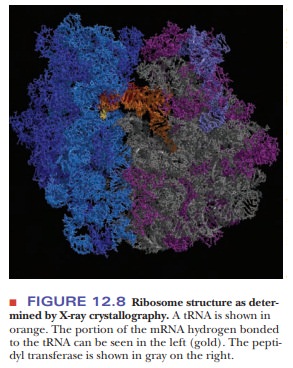
In Figure 12.8, a tRNA molecule (shown in orange) is base pairing with part of
the mRNA (gold) on the left. The tRNA extends into the peptidyltransferase
center on the right. Elucidation of the details of ribosomal structure is a
recent triumph of X-ray crystallography.
Chain Initiation
In all organisms, the synthesis of polypeptide
chains starts at the N-terminal end; the chain grows from the N-terminal end to
the C-terminal end. This is one of the reasons that scientists chose to record
DNA sequences from 5' to 3' and to focus on the coding strand of DNA and the
mRNA. The coding strand sequences are read from 5' to 3', the mRNA is read from
5' to 3', and the proteins are built from the N-terminus to the C-terminus. In
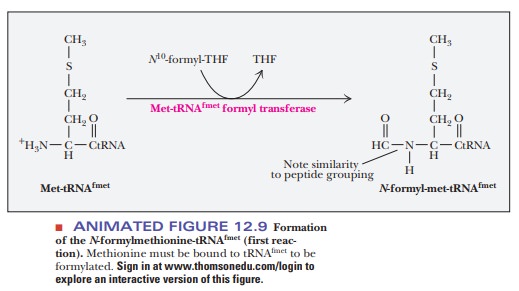
prokaryotes, the initial N-terminal amino acid of all proteins is N-formylmethionine (fmet) (Figure 12.9).
However, this residue often is removed by posttranslational processing after
the polypeptide chain is synthesized. There are two different tRNAs for
methionine in E. coli, one for
unmodified methionine and one for N-formylmethionine.
These two tRNAs are called tRNAmet and tRNAfmet,
respectively (the superscript identifies the tRNA). The aminoacyl-tRNAs that
they form with methionine are called met-tRNAmet and met-tRNAfmet,
respectively (the prefix identifies the bound amino acid). In the case of
met-tRNAfmet, a formylation reaction takes place after methionine is
bonded to the tRNA, producing N-formylmethionine-tRNAfmet
(fmet-tRNAfmet). The source of the formyl group is N10-formyltetrahydrofolate.
Methionine bound to tRNAmet is not formylated.
Both tRNAs (tRNAmet and tRNAfmet)
contain a specific sequence of three bases (a triplet), 3'-UAC-5', which
base-pairs with the sequence 5'-AUG-3'
in the mRNA sequence. The tRNAfmet triplet in question, 3'-UAC-5',
recognizes the AUG triplet, which is the start
signal when it occurs at the beginning of the mRNA sequence that directs
the synthesis of the polypeptide. The same 3'-UAC-5' triplet in tRNAmet
recognizes the AUG triplet when it is found in an internal position in the mRNA
sequence.
The start of polypeptide synthesis requires the formation of an initiationcomplex (Figure 12.10). At least eight components enter into the formationof the initiation complex, including mRNA, the 30S ribosomal subunit, fmet-tRNAfmet, GTP, and three protein initiation factors, called IF-1, IF-2, and IF-3. The IF-3 protein facilitates the binding of mRNA to the 30S ribosomal subunit. It also appears to prevent premature binding of the 50S subunit, which takes place in a subsequent step of the initiation process. IF-2 binds GTP and aids in the selection of the initiator tRNA (fmet-tRNAfmet) from all the other aminoac-ylated tRNAs available. The function of IF-1 is less clear; it appears to bind to IF-3 and to IF-2, and it facilitates the action of both. It also catalyzes the sepa-ration of the 30S and the 50S ribosomal subunits being recycled for another round of translation. The resulting combination of mRNA, the 30S ribosomal subunit, and fmet-tRNAfmet is the 30S initiation complex (Figure 12.10). A 50S ribosomal subunit binds to the 30S initiation complex to produce the 70Sinitiation complex. The hydrolysis of GTP to GDP and Pifavors the processby providing energy; the initiation factors are released at the same time.
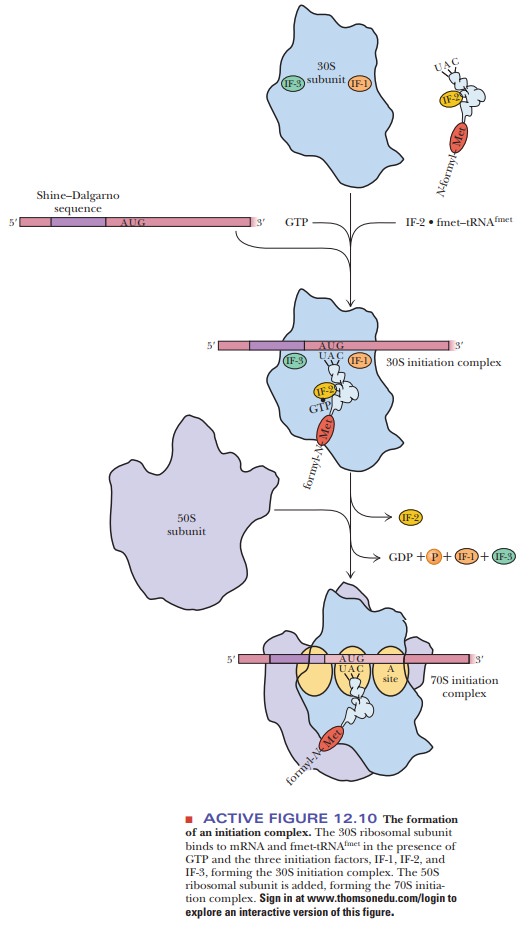
The correct
positioning of the initiator tRNA is maintained as a result of a small
difference between it and tRNA for an internal methionine. A single C–A
mis-matched base pair near the acceptor stem allows the 30S subunit to
recognize the initiator tRNA.
How does the ribosome know where to start translating?
For the
mRNA to be translated correctly, the ribosome must be placed at the correct
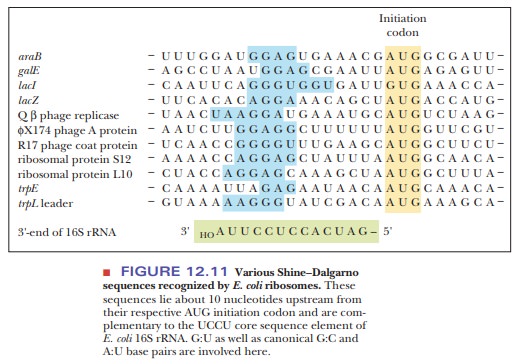
start location. The start signal is preceded by a purine-rich leader segment of
mRNA, called the Shine–Dalgarno sequence
(5'-GGAGGU-3') (Figure 12.10), which usually lies about 10 nucleotides upstream
of the AUG start signal (also known as the initiation codon) and acts as a
ribosomal binding site. Figure 12.11 gives some characteristic Shine–Dalgarno
sequences. This purine-rich area binds to a pyrimidine-rich sequence on the 16S
ribosomal RNA part of the 30S subunit and aligns it for proper translation
beginning with the AUG start codon.
Chain Elongation
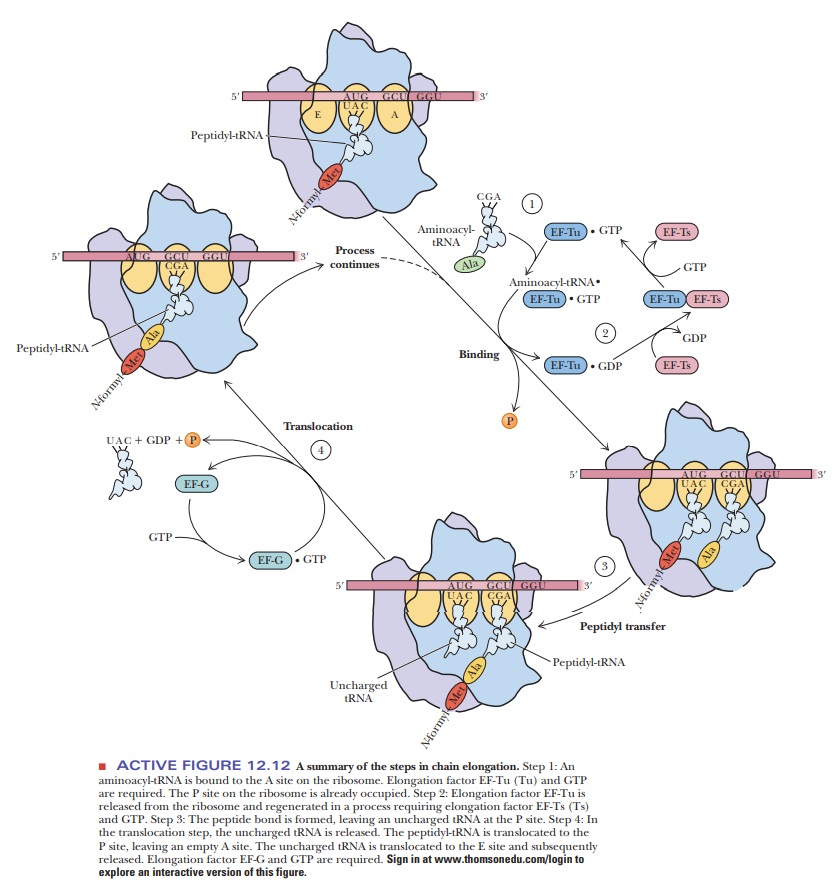
The
elongation phase of prokaryotic protein synthesis (Figure 12.12) uses the fact
that three binding sites for tRNA are present on the 50S subunit of the 70S
ribosome. The three tRNA binding sites are called the P (peptidyl) site, the A(aminoacyl)
site, and the E (exit) site. The
P site binds a tRNA that carries apeptide chain, and the A site binds an
incoming aminoacyl-tRNA. The E site carries an uncharged tRNA that is about to
be released from the ribosome. Chain elongation begins with the addition of the
second amino acid specified by the mRNA to the 70S initiation complex (Step 1).
The P site on the ribosome is the one initially occupied by the fmet-tRNAfmet in the
70S initiation complex. The second aminoacyl-tRNA binds at the A site. A
triplet of tRNA bases (the anticodon AGC in our example) forms hydrogen bonds
with a triplet of mRNA bases (GCU, the codon for alanine, in this example). In
addition, GTP and two protein elongation factors, EF-Tu and EF-Ts
(temperature-unstable and temperature-stable elongation factors, respectively),
are required (Step 2). EF-Tu guides the aminoacyl-tRNA into part of the A site
and aligns the anticodon with the mRNA codon. Only when the match is found to
be correct is the aminoacyl-tRNA inserted completely into the A site. GTP is
hydrolyzed and EF-Tu dissociates. EF-Ts is involved in regeneration of
EF-Tu-GTP. This small EF-Tu protein (43 kDa) is the most abundant protein in E. coli, comprising 5% of the dry weight
of the cell.
Why is EF-Tu so important in E. coli?
It has recently been shown that EF-Tu is involved in another level of translation fidelity. When the correct amino acid is bound to the correct tRNA, EF-Tu is efficient at delivering the activated tRNA to the ribosome. If the tRNA and amino acid are mismatched, then either the EF-Tu does not bind the activated tRNA very well, in which case it does not deliver it well to the ribosome, or it binds the activated tRNA too well, in which case it does not release it from the ribosome.
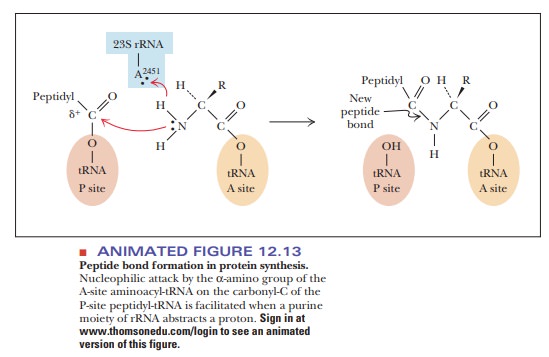
A peptide bond is then formed in a
reaction catalyzed by peptidyl
transferase, which is a part of the 50S subunit (Step 3). The mechanism for
this reaction is shown in Figure 12.13. The α-amino group of the amino acid in the A site
per-forms a nucleophilic attack on the carbonyl group of the amino acid linked
to the tRNA in the P site. There is now a dipeptidyl-tRNA at the A site and a
tRNA with no amino acid attached (an “uncharged
tRNA” ) at the P site.
A translocation step then takes place
before another amino acid can be added to the growing chain (Figure 12.12, Step
4). In the process, the uncharged tRNA moves from the P site to the E site,
from which it is subse-quently released; the peptidyl-tRNA moves from the A
site to the vacated P site. In addition, the mRNA moves with respect to the
ribosome. Another elonga-tion factor, EF-G, also a protein, is required at this
point, and once again GTP is hydrolyzed to GDP and Pi.
The
three steps of the chain elongation process are aminoacyl-tRNA bind-ing,
peptide bond formation, and translocation (Steps 1, 3, and 4 in Figure 12.12).
They are repeated for each amino acid specified by the genetic message of the
mRNA until the stop signal is reached. Step 2 in Figure 12.12 shows the
regeneration of aminoacyl-tRNA.
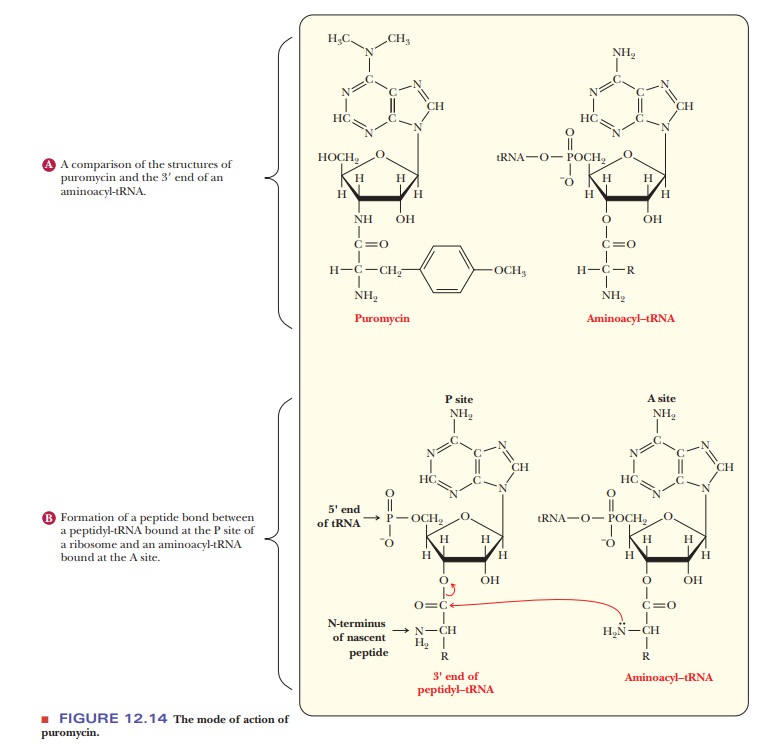
Much of
the information about this phase of protein synthesis has been gained from the
use of inhibitors. Puromycin is a structural analog for the 3' end of an
aminoacyl-tRNA, making it a useful probe to study chain elonga-tion (Figure
12.14). In an experiment of this sort, puromycin binds to the A site, and a
peptide bond is formed between the C-terminus of the growing polypeptide and
the puromycin. The peptidyl puromycin is weakly bound to the ribosome and
dissociates from it easily, resulting in premature termina-tion and a defective
protein. Puromycin also binds to the P site and blocks the translocation
process, although it does not react with peptidyl-tRNA in this case. The
existence of A and P sites was determined by these experiments with puromycin.
Chain Termination
A stop signal is required for the termination of protein synthesis. The codons UAA, UAG, and UGA are the stop signals. These codons are not recognized by any tRNAs, but they are recognized by proteins called release factors (Figure 12.15). One of two protein release factors (RF-1 or RF-2) is required, as is GTP, which is bound to a third release factor, RF-3. RF-1 binds to UAA and UAG, and RF-2 binds to UAA and UGA. RF-3 does not bind to any codon, but it does facilitate the activity of the other two release factors. Either RF-1 or RF-2 is bound near the A site of the ribosome when one of the termination codons is reached. The release factor not only blocks the binding of a new aminoacyl-tRNA but also affects the activity of the peptidyl transferase so that the bond between the carboxyl end of the peptide and the tRNA is hydrolyzed.
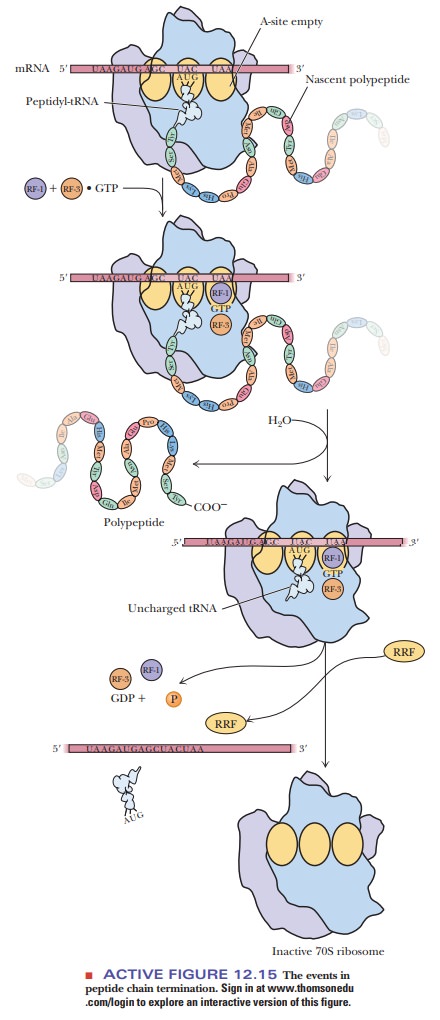
GTP is hydrolyzed
in the process. The whole complex dissociates, setting free the release
factors, tRNA, mRNA, and the 30S and 50S ribosomal subunits. All these
components can be reused in further protein synthesis. Table 12.3 summarizes
the steps in protein synthesis and the components required for each step. The
following Biochemical Connections box describes an interesting variation on
stop codons.
The Ribosome Is a Ribozyme
Until recently, proteins were thought to be the only molecules with catalytic ability. Then the self-splicing ability of the Tetrahymena snRNP showed that RNA can also catalyze reactions. In 2000, the complete structure of the large ribosomal subunit was determined by X-ray crystallography to 2.4-Ă… (0.24-nm) resolution (Figure 12.16).
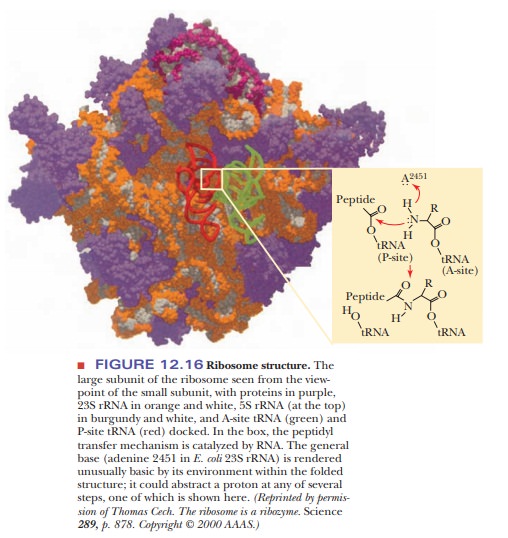
Ribosomes had been studied for 40 years, but the complete structure had
been elusive. When the active sites for peptidyl transferase were looked at, it
turned out that there is no protein in the vicinity of the new peptide bond,
showing once again that RNA has catalytic ability. This is an exciting finding
because it answers questions that have been plaguing scientists for decades. It
was assumed that RNA was the first genetic material, and RNA can encode
proteins that act as catalysts; but, because it takes proteins to do the
translation, how could the first proteins have been created? With the discovery
of an RNA-based peptidyl transferase, it was suddenly possible to imagine an
“RNA world” in which the RNA both carried the message and processed it. This discovery
is very intriguing, but it has not yet been accepted by many researchers, and
some evidence questions the nature of catalytic RNA. One study showed that
mutations of the putative RNA bases involved in the catalytic mechanism do not
significantly reduce the efficiency of peptidyl transferase, throwing into
question whether the RNA is chemically involved in the catalysis.
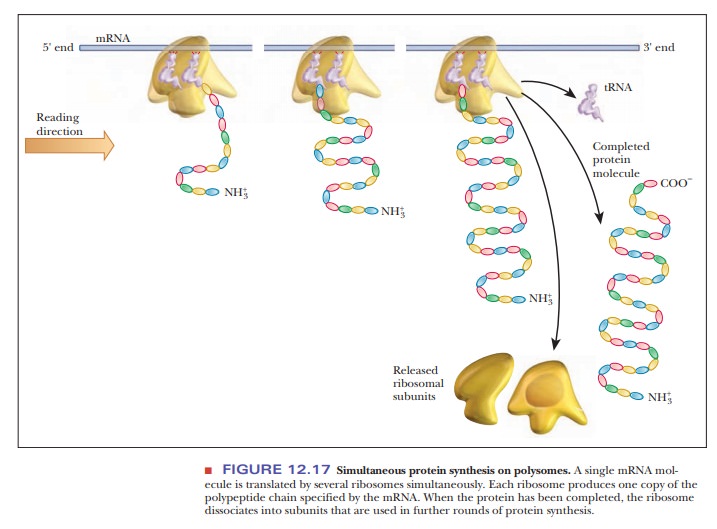
Polysomes
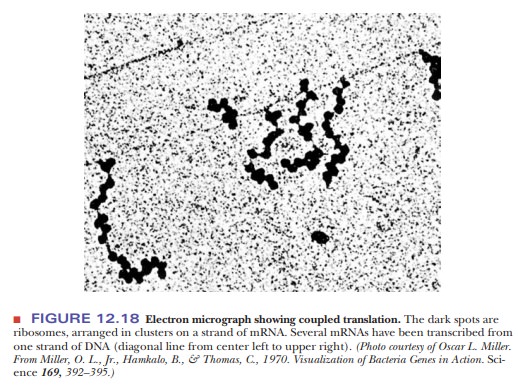
In our description of protein synthesis, we have considered, up to now, the reactions that take place at one ribosome. It is, however, not only possible but quite usual for several ribosomes to be attached to the same mRNA. Each of these ribosomes bears a polypeptide in one of various stages of completion, depending on the position of the ribosome as it moves along the mRNA (Figure 12.17). This complex of mRNA with several ribosomes is called a polysome; an alternative name is polyribosome. In prokaryotes, translation begins very soon after mRNA transcription. It is possible for a molecule of mRNA that is still being transcribed to have a number of ribosomes attached to it that are in various stages of translating that mRNA. It is also possible for DNA to be in various stages of being transcribed. In this situation, several molecules of RNA polymerase are attached to a single gene, giving rise to several mRNA molecules, each of which has a number of ribosomes attached to it. The prokaryotic gene is being simultaneously transcribed and translated. This process, which is called coupled translation (Figure 12.18), is possible in prokaryotes because of the lackof cell compartmentalization. In eukaryotes, mRNA is produced in the nucleus, and most protein synthesis takes place in the cytosol.
Summary
The unique and elegant structure of the
ribosome allows the binding of aminoacyl-tRNA molecules and mRNA. The ribosome
catalyzes the nucleophilic attack of one amino acid on the next, allowing for
protein synthesis.
Protein synthesis begins at an AUG codon on the
mRNA. The ribosome and mRNA forms an initiation complex that includes the two
main ribo-somal subunits, the mRNA, GTP, and three initiation factors, IF1,
IF2, and IF3.
The ribosome locates the correct AUG to start translation by
binding to a consensus sequence called the Shine–Dalgarno sequence.
The first aminoacyl-tRNA bound to the ribosome carries N-formylmethio-nine, and it is initially
bound to the P site of the ribosome.
In chain elongation, the second amino acyl-tRNA
binds to the A site. This amino acid’s α-amino group performs a nucleophilic attack on
the carbonyl group of the N-formylmethionine
in the peptidyl transfer reac-tion. In a translocation step, the ribosome then moves
one codon, leaving a dipeptidyl-tRNA in the A site and moving the uncharged
tRNA to the exit site. The process continues with a new aminoacyl-tRNA entering
the P site. The uncharged tRNA is then ejected from the E site.
When the ribosome encounters a stop codon, the chain is terminated
in a process requiring GTP and three protein release factors.
The ribosome is actually a ribozyme. There are
no amino acids at the active site where the peptidyl transferase reaction
occurs. Specific bases on the rRNA are believed to catalyze the reaction.
Related Topics