Chapter: Biochemistry: Protein Synthesis: Translation of the Genetic Message
Codon–Anticodon Pairing and Wobble
Codon–Anticodon Pairing and
Wobble
A codon
forms base pairs with a complementary anticodon
of a tRNA when an amino acid is incorporated during protein synthesis. Because
there are 64 possible codons, one might expect to find 64 types of tRNA but, in
fact, the number is less than 64 in all cells.
If there are 64 codons, how can there be less than 64 tRNA molecules?
Some
tRNAs bond to one codon exclusively, but many of them can recognize more than
one codon because of variations in the allowed pattern of hydrogen bonding.
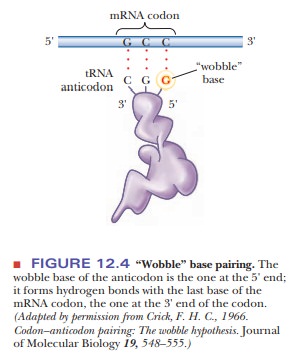
This variation is called “wobble”
(Figure 12.4), and it applies to the first base of an anticodon, the one at the
5' end, but not to the second or the third base. Recall that mRNA is read from
the 5' to the 3' end. The first (wobble) base of the anticodon hydrogen-bonds
to the third base of the codon, the one at the 3' end. The base in the wobble
position of the anticodon can base-pair with several different bases in the
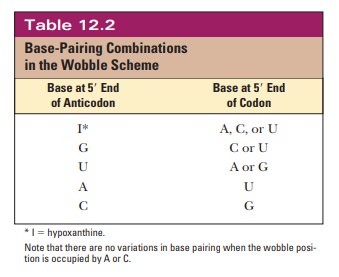
codon, not just the base specified by Watson–Crick base pairing (Table 12.2).
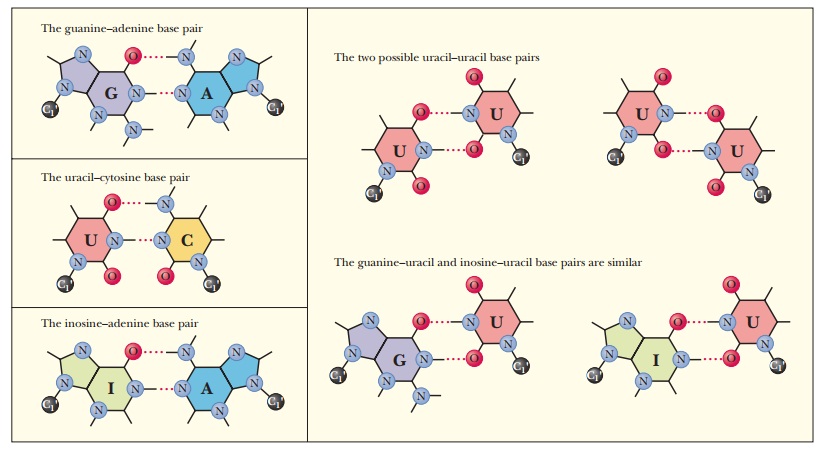
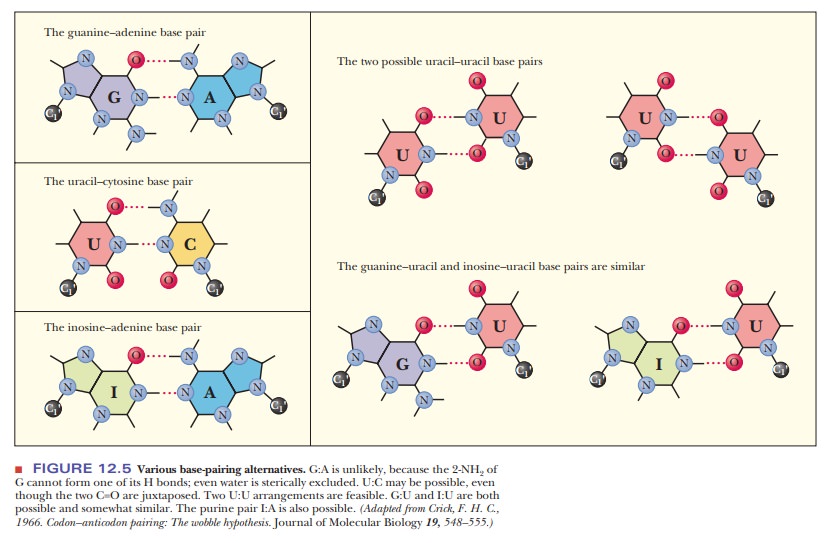
When the
wobble base of the anticodon is uracil, it can base-pair not only with adenine,
as expected, but also with guanine, the other purine base. When the wobble base
is guanine, it can base-pair with cytosine, as expected, and also with uracil,
the other pyrimidine base. The purine base hypoxanthine fre-quently occurs in
the wobble position in many tRNAs, and it can base-pair with adenine, cytosine,
and uracil in the codon (Figure 12.5). Adenine and cyto-sine do not form any
base pairs other than the expected ones with uracil and guanine, respectively
(Table 12.2). To summarize, when the wobble position is occupied by I (from
inosine, the nucleoside made up of ribose and hypoxan-thine), G, or U,
variations in hydrogen bonding are allowed; when the wobble position is
occupied by A or C, these variations do not occur.
The
wobble model provides insight into some aspects of the degeneracy of the code.
In many cases, the degenerate codons for a given amino acid differ in the third
base, the one that pairs with the wobble base of the anticodon. Fewer different
tRNAs are needed because a given tRNA can base-pair with several codons. As a
result, a cell would have to invest less energy in the synthesis of needed
tRNAs. The existence of wobble also minimizes the damage that can be caused by
misreading of the code. If, for example, a leucine codon, CUU, were to be
misread as CUC, CUA, or CUG during transcription of mRNA, this codon would
still be translated as leucine during protein synthesis; no damage to the
organism would occur. We saw in earlier that drastic consequences can result
from misreading the genetic code in other codon positions, but here we see that
such effects are not inevitable.
A universal code is one that is the same in all organisms. The universality of the code has been observed in viruses, prokaryotes, and eukaryotes. However, there are some exceptions.
Some codons seen in mitochondria are different from
those seen in the nucleus. There are also at least 16 organisms that have code
variations. For example, the marine alga Acetabularia
translates the stan-dard stop codons, UAG and UAA, as a glycine rather than as
a stop. Fungi of the genus Candida
translate the codon CUG as a serine, where that codon would specify leucine in
most organisms. The evolutionary origin of these differences is not known at
this writing, but many researchers believe that understanding these code
variations is important to understanding evolution.
Related Topics