Chapter: Microbiology and Immunology: Genetic Engineering and Molecular Methods
Polymerase Chain Reaction
Polymerase Chain Reaction
In 1983, Kary Mullis developed a new technique that made it possible to synthesize large quantities of a DNA fragment without cloning it. This technique is called polymerase chain reaction (PCR) and has great practical importance and impact on biotechnology. By this technique, large quantities of a particular DNA sequence can be prepared.
In PCR,
oligonucleotide sequences identical to those flanking the targeted sequence are
first synthesized. These synthetic oligonucleotides are usually about 20
nucleotides long and serve as primers for DNA synthesis. Pieces ranging in size
from less than 100 bp to several 1000 bp in length can be amplified, and only
10–100 pmol primer is required. The concentration of target DNA can be as low
as 10–15 mL.
The
reaction mix for PCR contains (a) the
target DNA (b) a very large excess of
the desired primers, (c) a
thermostable DNA polymerase, and (d )
four deoxyribonucleoside triphosphates. Only DNA polymerases that are able to
function at the high temperatures can be employed in the PCR technique. aq poly-merase from the thermophilic
bacterium Thermus aquaticus andthe Vent polymerase from Thermococcus litoralis are the two
popu-lar enzymes used in the PCR.
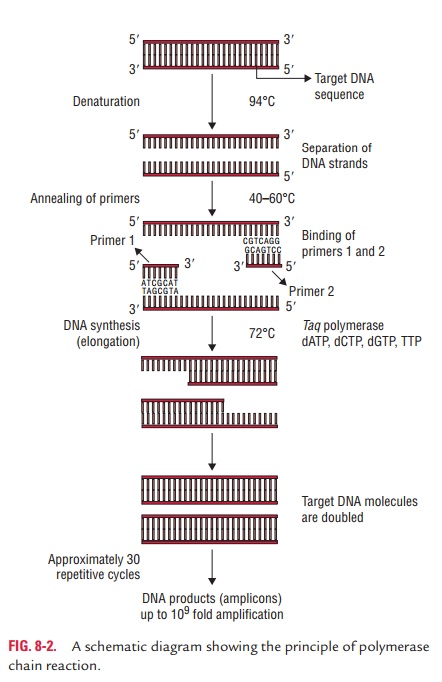
The PCR
cycle takes place in three steps as follows (Fig. 8-2):
Step 1: The target DNA containing the sequence to be ampli-fied is heat
denatured to separate its complementary strands. Normally the target DNA is
between 100 and 5000 bp in length.
Step 2: The temperature is lowered so that the primers cananneal to the DNA
on both sides of the target sequence. Because the primers are present in
excess, the targeted DNA strands normally anneal to the primers rather than to
each other.
Step 3: DNA polymerase extends the primers and synthe-sizes copies of the
target DNA sequence using the deoxyri-bonucleoside triphosphates.
At the
end of one cycle, the targeted sequences on both strands are copied. When the
three-step cycle is repeated, the four strands from the first cycle are copied
to produce eight fragments. The third cycle yields 16 products. Theoretically,
20 cycles will produce about one million copies of the target DNA sequence, and
30 cycles yield around one billion copies.
The PCR
technique has now been automated and is carried out through a specially
designed machine called thermocy-cler. Currently, a thermocycler or PCR machine
can carry out 25 cycles and amplify DNA 105 times in as little as 57
minutes. During a typical cycle, the DNA is denatured at 94°C for 15 seconds;
then the primers are annealed and extended (steps 2 and 3) at 68°C for 60
seconds. PCR technology is improving continually and undergoing many changes as
follows:
a) Nowadays, RNA can be efficiently used in PCR proce-dures. The Tth DNA polymerase, a recombinant Thermus thermophilus DNA polymerase, will transcribe RNA to DNAand then amplify the DNA. Cellular RNAs and RNA viruses may be studied even when the RNA is present in very small amounts (as few as 100 copies can be transcribed and amplified).
b)
Also,
PCR can quantitate DNA products without the use of isotopes. This allows one to
find the initial amount of target DNA in less than an hour using automated
equip-ment. Quantitative PCR is quite valuable in virology and gene expression
studies.
c)
As
mentioned earlier, the target DNA to be amplified is normally less than about
5000 bp in length. A long PCR technique has been developed that will amplify
sequences up to 42 kilo bases long. It depends on the use of error-correcting
polymerases because Taq polymerase is
error-prone.
d) Multiplex
PCR is another modification of PCR in which twoor
more target sequences can be demonstrated simultane-ously in a single specimen
at the same time. This method uses two or more primer sets designed for
amplification of different targets. Multiplex is now increasingly evaluated for
simultaneous demonstration of two or more patho-gens in a clinical specimen.
e)
Real-time PCR is the most recent development. It is so
named,because the PCR amplicons can be detected in real time. In fact, “real
time” refers to the detection of amplicons after each PCR cycle. Several
commercial instruments are available that combine PCR amplification of target
DNA with detection of amplicons in the same closed vessel. Probe detection
formats involve detecting fluorophores. Results are semiquantitative and can be
obtained in considerably less time than it takes to perform a conven-tional PCR
assay.
Related Topics