Chapter: Biotechnology Applying the Genetic Revolution: Environmental Biotechnology
Function or Activity Based Evaluation of the Environment
FUNCTION-
OR ACTIVITY-BASED EVALUATION OF THE ENVIRONMENT
Besides these sequence-based
approaches, metagenomic libraries can be screened for various functions (Fig.
12.6). Functional approaches include expression screening with various genetic
traps and phage biopanning. Even stable isotope probing (see earlier
discussion) could be categorized as a function-based approach if the labeled
substrate is a specific metabolite that enriches the culture based on metabolic
function. Screening a metagenomic library by sequencing has its limits. The
function of many genes from exotic organisms cannot be identified by their
sequence. In addition, using known genes to screen for new members of a gene
family might miss an entire novel class of genes. For example, if a researcher
were trying to find enzymes similar to bacteriorhodopsin, primers similar to
known bacteriorhodopsins would be used to screen the library. Some genes would
be identified, but others would be missed if their sequences were too
divergent. Screening this library for light-driven proton pumping would
identify any enzyme that carried out this function, regardless of its sequence.
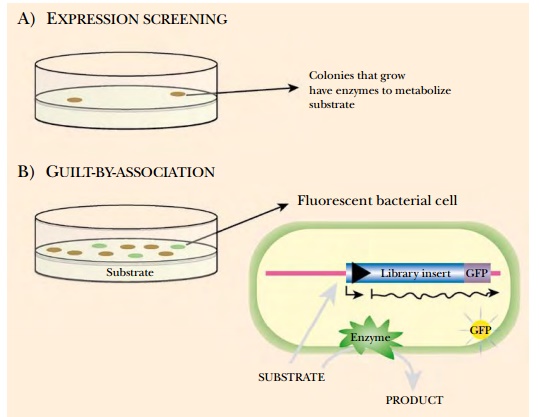
Expression screening depends
on the choice of an expression vector. Proteins are expressed from the
metagenomic DNA fragments when they are cloned into a vector with
transcriptional and translational start and stop sequences. Then an easy assay
for the target function must be devised. For example, the library clones might
be plated on a particular toxic pollutant. If any library inserts encoded an
enzyme that metabolized the pollutant, that particular library clone would
grow. The DNA insert is then isolated and identified by sequencing.
Another functional screen
involves fusing the metagenomic DNA in frame with a promoterless gene for green
fluorescent protein (GFP). The genes for many enzymes are turned on by their
own substrates. Therefore, if this type of gene is cloned in front of
promoterless GFP, the GFP gene will be regulated by the same substrate. If the
substrate of interest is included in the growth medium for the library, any
clone with genes that are activated by the substrate will also produce GFP.
These cells will fluoresce green. FACS
provides a quick and easy way to isolate the fluorescent clones.
The main barrier to
function-based analyses is successful gene expression. Getting a library host
such as E. coli to express foreign
genes is hit or miss, because some may be toxic to the host, some may require
other factors for expression, and others may have very low activity. Another
problem is simply volume. The number of potential clones that have any chosen gene
of interest is usually low, and excessive numbers of clones must be screened in
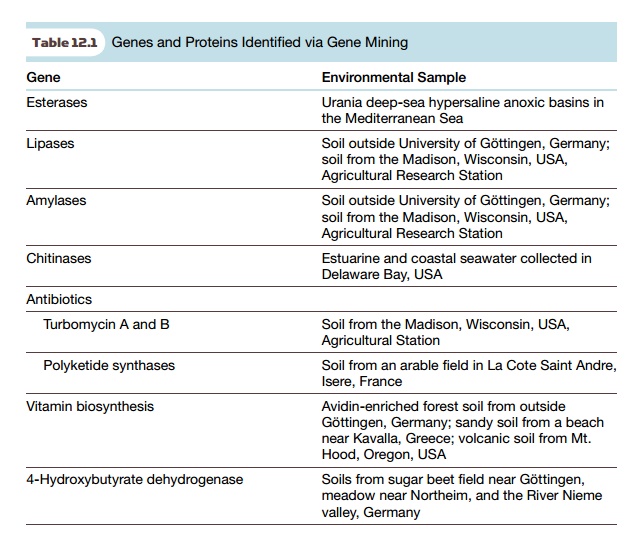
order to identify just one or two genes. For example, the lipases identified
from German soil (see Table 12.1) were only found in 1 of 730,000 different
metagenomic clones. In another example, only two novel Na+/H+
antiporters were found after screening 1,480,000 clones. This is why culture
enrichment strategies are an important aspect of creating metagenomic libraries
(see earlier discussion).
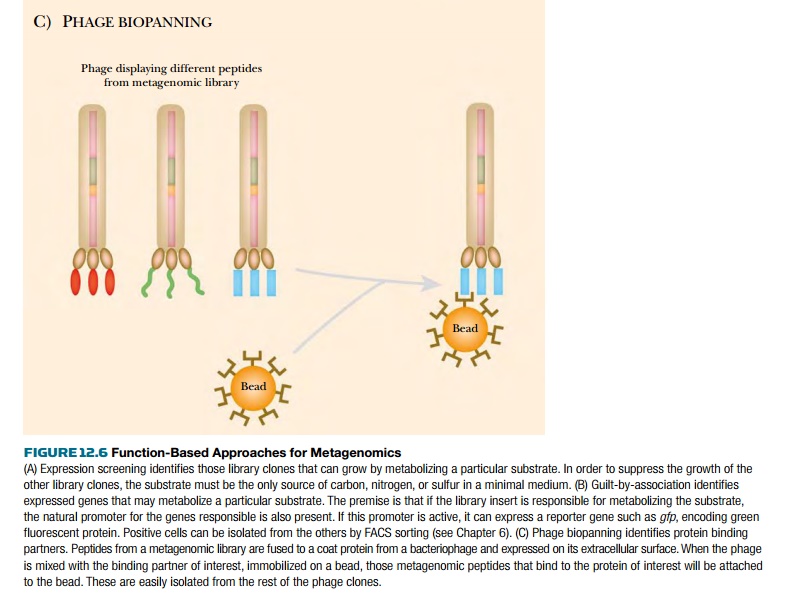
Metagenomic phage biopanning
uses basically the same method as phage display. Cloned DNA inserts are
expressed as fusion proteins with a phage coat protein and displayed on the
phage surface. Because the displayed proteins carry only segments of foreign
protein, the problems associated with heterologous expression are lessened. The
cloned DNA is from any organisms found within the environmental sample. Thus
the expressed protein segments could be any part of an enzyme, a membrane
protein, etc.; therefore, the success with a phage display library relies on
the screening method. For example, phage biopanning can identify binding
partners for a particular pollutant, metabolite, or even another protein. If
the target molecule is immobilized on a bead, then any phage carrying a protein
segment that binds the target will stick to the bead. The phage can then be
isolated, and the DNA insert sequenced to identify the sequence responsible.
Related Topics