Chapter: Biotechnology Applying the Genetic Revolution: Environmental Biotechnology
Culture Enrichment for Environmental Samples
CULTURE
ENRICHMENT FOR ENVIRONMENTAL SAMPLES
Various methods are used to
enhance the starting material for metagenomics research, because a metagenomic
library is only as good as its contents. Enrichment strategies include stable
isotope probing (SIP), BrdU enrichment, and suppressive subtraction
hybridization.
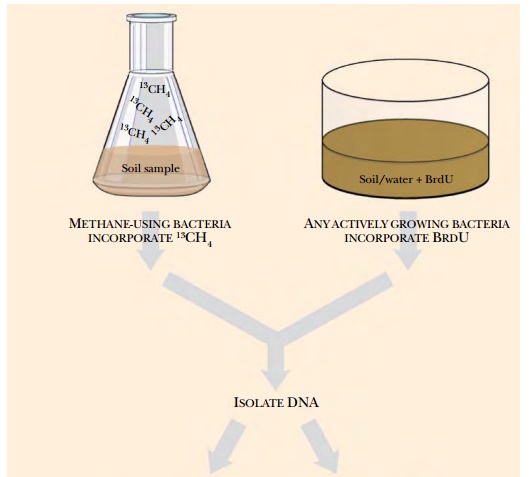
Stable isotope probing (SIP) was originally developed to trace single carbon compounds during their metabolism by cultured methylotrophs (bacteria specialized for growth on single-carbon compounds). Labeled precursor carbons were traced into fatty acids during bacterial growth. The method was adapted to the environmental samples used to create metagenomic libraries. Here, an environmental sample of water or soil is first mixed with a precursor such as methanol, phenol, carbonate, or ammonia, that has been labeled with a stable isotope such as 15N, 13C, or 18O (Fig. 12.2). If the organisms in the sample metabolize the precursor substrate, the stable isotope is incorporated into their genome. Then, when the DNA from the sample is isolated and separated by centrifugation, the genomes that incorporated the labeled substrate will be “heavier” and can be separated from the other DNA in the sample. The heavier DNA will migrate further in a cesium chloride gradient during centrifugation. As described later, the DNA can either be used directly or cloned into vectors to make a metagenomic library. This technique is particularly useful to find new organisms that can degrade contaminants, such as phenol in the example given.
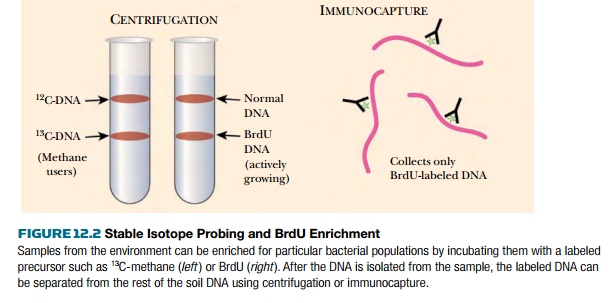
Adding 5-bromo-2-deoxyuridine
(BrdU) is a related technique for enriching for the DNA of active bacteria in
an environmental sample. Rather than entering only a metabolic subset of
bacteria, BrdU is incorporated into the DNA and RNA of any actively growing
bacteria or viruses. Note that bacteria and viruses that are dormant or dead,
as well as free DNA, will not be labeled by this method. As before, the soil or
water is isolated and incubated with BrdU.
Any bacteria that are
actively growing will take up the nucleotide analog and incorporate it into its
DNA. Next, the BrdU-labeled DNA is isolated either with antibodies to BrdU or
by density gradient centrifugation (see Fig. 12.2).
RNA-SIP focuses on isolating
RNA from the environmental sample (rather than DNA). Small subunit ribosomal
RNA (SSU rRNA), that is, the 16S rRNA of bacteria or the 18S rRNA of
eukaryotes, is an excellent biomarker because it is essential to all cellular
life, it is very abundant within a cell, it is variable among different
species, and there is an enormous database of different SSU rRNA sequences
making identification relatively easy. In RNA SIP the SSU rRNA in the
environmental sample is labeled. As described earlier, 13C-labeled precursors
are supplied to the environmental sample. These are incorporated into SSU rRNA
independently of cell division because ribosomal RNA is produced in any cell
that is making proteins, not just cells undergoing replication. This technique
provides information on bacteria that are dormant as well as those that are
more active. Much as before, the RNA is isolated and separated on a gradient by
centrifugation. The rRNA bands tend to aggregate together during
centrifugation. Therefore, each fraction must be repeatedly separated from the
others. The final SSU rRNA fraction may still contain some contaminating
nonlabeled rRNAs, so the fraction must be evaluated with care.
RNA-SIP can be used to
identify a variety of microorganisms in environmental samples. For example, water
from an aerobic industrial wastewater plant was evaluated for phenol- degrading
microorganisms. The water was incubated with 13C-labeled phenol, and the SSU
rRNA was isolated by centrifugation. The rRNAs were isolated and amplified with
RT-PCR followed by denaturing gradient gel electrophoresis. The bands were
subjected to mass spectrometry to identify which rRNA sequence was most
abundant. Interestingly, an organism belonging to the genus Thauera in the
β-Proteobacteria was abundant, even though this organism was most usually found
in denitrifying conditions. It was previously thought that pseudomonads were
degrading the phenol.
Another culture-enrichment
technique, suppressive subtraction hybridization (SSH), takes advantage of the
genetic differences between samples from two different areas. During standard
subtractive hybridization, two different samples are hybridized and the mRNA that
is the same is removed, leaving only mRNA that is different between the two
samples. SSH works by the same principle. First, two different conditions must
be established. For example, one soil sample from a polluted site could be
compared with nearby soil that is not contaminated. The two soil samples will
differ in their content of microorganisms, and those microorganisms enriched in
the contaminated site could potentially metabolize the contaminant.
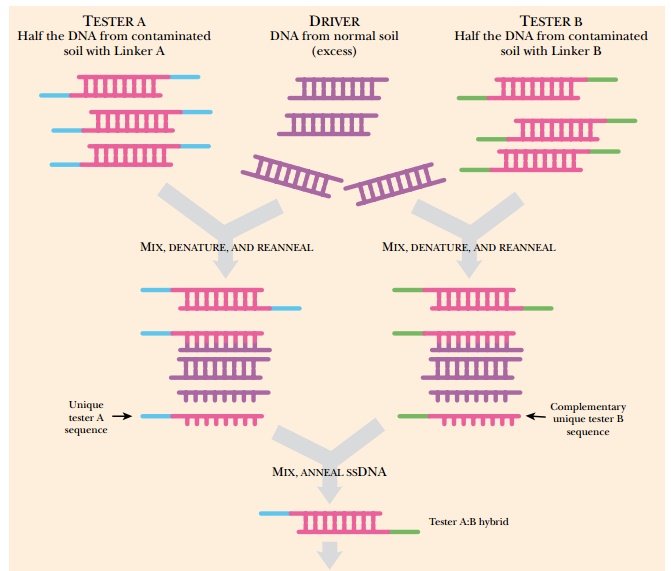
When DNA from each sample is
isolated, the contaminated soil is considered the tester DNA and the “normal”
soil is the driver sample. The tester sample is divided into two, and two
different linkers are added to the ends of the DNA to form tester A and tester
B. Tester A (with linker A), tester B (with linker B), and driver DNA are all
mixed, denatured to make them single-stranded, and then rehybridized. The
driver DNA is in excess to the testers, which ensures that DNA fragments from
bacteria outside the contamination site outnumber those from the contaminated
site. The driver DNA will anneal to all the common DNA fragments, making these
double-stranded and with only one strand connected to the linker. All the
tester DNA that is unique and not found in the uncontaminated soil will be free
to hybridize with itself, forming A:A, B:B, or A:B hybrids. PCR primers are
added to the hybridization mix; one primer recognizes linker A and the other
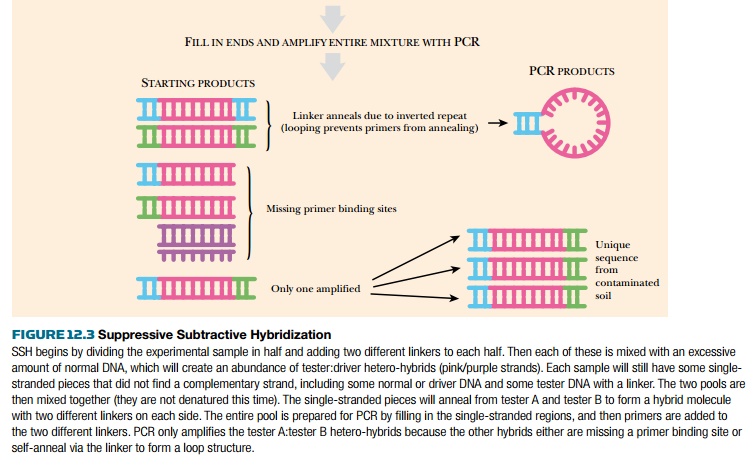
primer is for linker B. As shown in Fig. 12.3, PCR will amplify only those
hybrids that are tester:tester. Furthermore, because the A:A and B:B hybrids
have inverted linkers, these hybrids will form a “panlike” structure during
annealing and will not be amplified by PCR. Thus only A:B hybrids are
amplified, and these represent unique sequences found only in the contaminated site.
Related Topics