Chapter: Biotechnology Applying the Genetic Revolution: Forensic Molecular Biology
Using the Polymerase Chain Reaction (PCR)
USING
THE POLYMERASE CHAIN REACTION (PCR)
The polymerase chain reaction (PCR) is a procedure for amplifying tiny amounts of DNA and is used when there is too little DNA, or the DNA is too degraded for DNA fingerprinting via the RFLP approach. The details of PCR have already been discussed. PCR machines can amplify a segment of DNA (100 to 3000 bp long) in a few hours, starting from only a picogram (10 –12 g), although microgram (10 –6 g) quantities or larger are better. In fact, PCR can be used successfully to analyze DNA from a single cell.
Whereas classical DNA fingerprinting
requires relatively long strands of DNA, PCR can be used on short segments of
DNA. PCR is most useful for regions of the DNA with high individual
variability. Small regions with high person-to-person variability are the best
to amplify. If the sequences of two samples match in several highly variable
regions, they are probably from the same person. In current practice, forensic
DNA analysis is almost all done by PCR-based methodology.
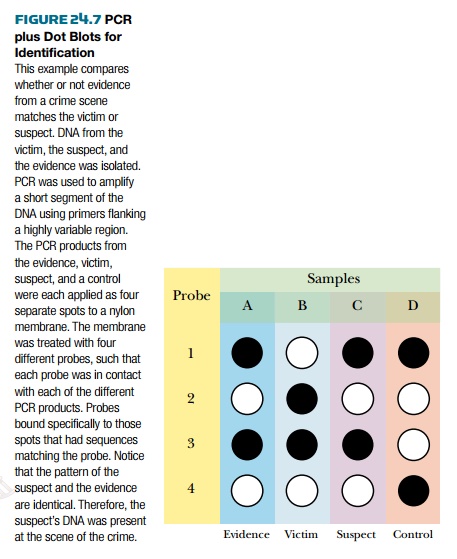
Once the DNA from the forensic sample has
been amplified by PCR, it is compared with DNA from the suspect, or suspects. Spots of both
DNA samples are bound to a membrane and tested for binding to a DNA probe that
is either radioactive or tagged with a fluorescent dye. The probe either binds
or doesn’t bind, so any spot is either positive or negative. This kind of test
is known as a dot blot ( Fig. 24.7 ). Thus, the major difference between PCR
and DNA fingerprinting is that DNA fingerprinting looks for differences in
fragment sizes, whereas PCR tests for the presence or absence of specific
stretches of DNA with identical (or almost identical) sequences. If necessary, segments
of DNA that have been amplified by PCR can be fully sequenced. In this case, we
do not need to rely just on hybridization as an indication of related
sequences. In the future, such sequence identification will most likely be done
by DNA array technology, which allows simultaneous analysis of multiple short
sequences.
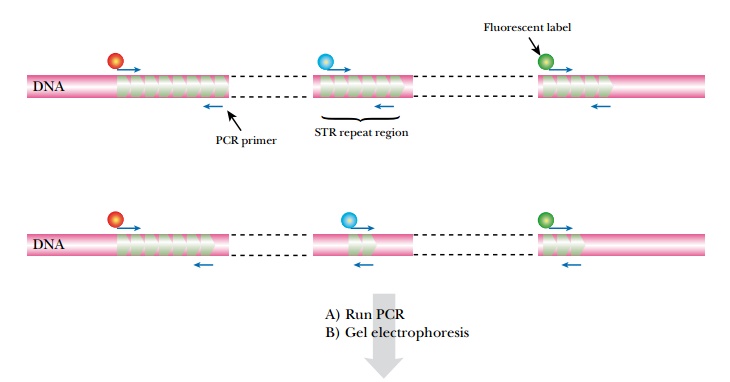
It is possible to analyze several STR
loci simultaneously by running several PCR reactions in the same tube using
different primers ( multiplex PCR ; Fig. 24.8 ). This requires that the multiple
sets of primers do not interfere with each other, which is often difficult to
achieve when running six or more amplifications together. Nonetheless, commercial
kits are now available that can run up to 13 STR analyses in one reaction tube.
Such a multiplex analysis gives a gel track with up to 2 N bands (where N
is the number of loci analyzed). For this example, 13 STR loci would
produce 26 bands. Fewer bands will be seen in individuals who are homozygous at
any of the chosen loci.
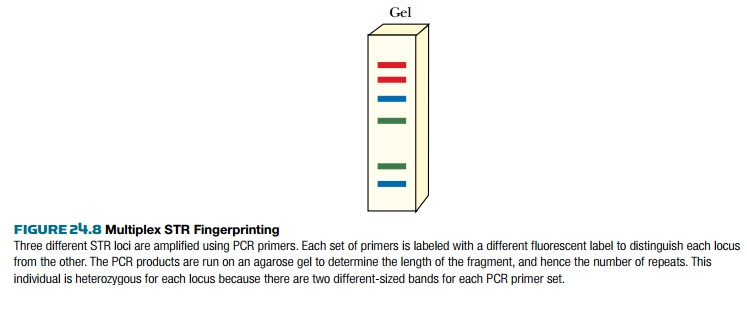
Despite the apparent complexity, the
STRs that are used derive from known sequences at known chromosomal locations,
and hence the individual bands can be identified and entered into computer
databases. Multiplex STR analysis is the basis of the national database set up
in the UK in 1995. Similar databases are now used in other European nations
and, since 1998, in the United States.
Related Topics