Chapter: Biochemistry: Protein Synthesis: Translation of the Genetic Message
The Genetic Code
The Genetic Code
Some of
the most important features of the code can be specified by saying that the
genetic message is contained in a triplet,
nonoverlapping, commaless, degenerate,universal code. Each of these terms
has a definite meaning that describes the wayin which the code is translated.
A triplet code means that a sequence of
three bases (called a codon) is
needed to specify one amino acid. The genetic code must translate the lan-guage
of DNA, which contains four bases, into the language of the 20 common amino
acids that are found in proteins. If there were a one-to-one relationship
between bases and amino acids, then the four bases could encode only four amino
acids, and all proteins would have to be combinations of these four. If it took
two bases to make a codon, then there would be 42
possible combina-tions of two bases for 16 possible amino acids, which is still
not enough. Thus, one could have guessed that a codon would have to be at least
three bases long. With three bases, there are 43
possibilities, or 64 possible codons, which is more than enough to encode the
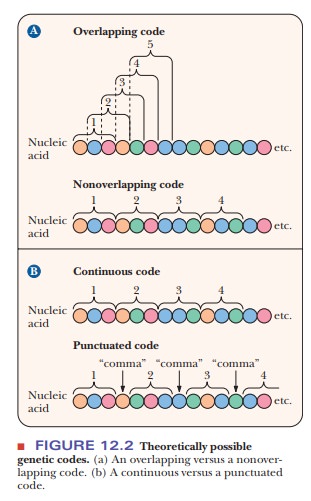
20 amino acids. The term nonoverlapping
indicates that no bases are shared between consecutive codons; the ribosome
moves along the mRNA three bases at a time rather than one or two at a time
(Figure 12.2). If the ribosome moved along the mRNA more than three bases at a
time, this situation would be referred to as “a punctuated code.” Because no
intervening bases exist between codons, the code is commaless. In a degener-ate code,
more than one triplet can encode the same amino acid. Sixty-four(4 X 4 X 4) possible triplets of the four bases occur
in RNA, and all are used to encode the 20 amino acids or one of the three stop
signals. Note that there is a big difference between a degenerate code and an
ambiguous one. Each amino acid may have more than one codon, so the genetic
code is a little redundant, but no codon can encode more than one amino acid.
If it did, the code would be ambiguous, and the protein-synthesizing machinery would
not know which amino acid should be inserted in the sequence. All 64 codons
have been assigned meanings, with 61 of them coding for amino acids and the
remaining - > serving as the termination signals (Table 12.1).
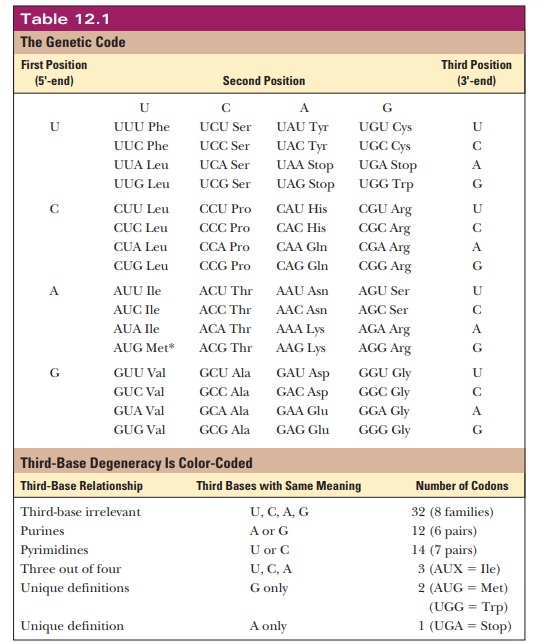
Two amino acids, tryptophan and methionine, have only one codon each, but the rest have more than one. A single amino acid can have as many as six codons, as is the case with leucine and arginine. Originally, the genetic code was thought to be a random selection of bases encoding amino acids. More recently, it is becoming clear why the code has withstood billions of years of natural selection. Multiple codons for a single amino acid are not randomly distributed in Table 12.1 but have one or two bases in common. The bases that are common to several codons are usually the first and second bases, with more room for variation in the third base, which is called the “wobble” base. The degeneracy of the code acts as a buffer against deleterious mutations. For example, for eight of the amino acids (L, V, S, P, T, A, G, and R), the third base is completely irrelevant. Thus, any mutation in the third base of these codons would not change the amino acid at that location. A mutation in the DNA that does not lead to a change in the amino acid translated is called a silent mutation.
In
addition, the second base of the codon also appears to be very important for
determining the type of amino acid. For example, when the second base is U, all
the amino acids generated from the codon possibilities are hydrophobic. Thus,
if the first or third base were mutated, the mutation would not be silent, but
the damage would not be as great because one hydrophobic amino acid would be
replaced with another. Codons sharing the same first letter often code for
amino acids that are products of one another or precursors of one another. A
recent paper in Scientific American
looked at the error rate for other hypothetical genetic codes and calculated
that, of 1 million possible genetic codes that could be conceived of, only 100
would have the effect of reducing errors in protein function when compared with
the real code. Indeed, it seems that the genetic code has withstood the test of
time because it is one of the best ways to protect an organism from DNA
mutations.
However,
to make matters more interesting, scientists have recently discov-ered that
some silent mutations are not as silent as they once thought.
How did scientists determine the genetic code?
The
assignment of triplets in the genetic code was based on several types of
experiments. One of the most significant experiments involved the use of
synthetic polyribonucleotides as messengers. When homopolynucleotides
(polyribonucleotides that contain only one type of base) are used as a syntheticmRNA for polypeptide synthesis
in laboratory systems, homopolypeptides(polypeptides that contain only one kind
of amino acid) are produced. When poly U is the messenger, the product is
polyphenylalanine. With poly A as the messenger, polylysine is formed. The
product for poly C is polyproline, and the product for poly G is polyglycine.
This procedure was used to establish the code for the four possible
homopolymers quickly. When an alternating copolymer (a polymer with an
alternating sequence of two bases) is the messenger, the product is an
alternating polypeptide (a polypeptide with an alternating sequence of two
amino acids). For example, when the sequence of the polynucleotide is
–ACACACACACACACACACACAC–, the polypeptide produced has alternating threonines
and histidines. There are two types of coding triplets in this polynucleotide,
ACA and CAC, but this experiment cannot establish which one codes for threonine
and which one codes for histidine. More information is needed for an
unambiguous assignment, but it is interesting that this result proves that the
code is a triplet code. If it were a doublet code, the product would be a
mixture of two homopolymers, one specified by the codon AC and the other by the
codon CA. (The terminology for the different ways of reading this message as a
doublet is to say that they have different reading
frames, /AC/ AC/ and /CA/CA/. In a triplet code, only one reading frame is
possible, namely, /ACA/CAC/ACA/CAC/, which gives rise to an alternating
polypeptide.) Use of other synthetic polynucleotides can yield other coding
assignments, but, as in our example here, many questions remain.
Other
methods are needed to answer the remaining questions about codon assignment.
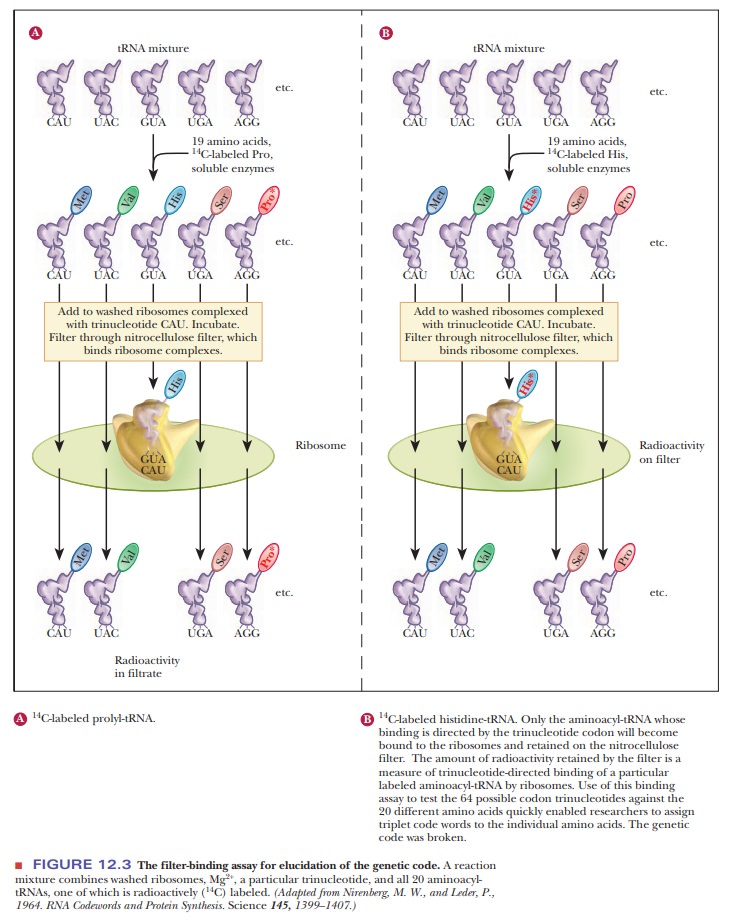
One of the most useful methods is the filter-binding
assay (Figure 12.3). In this technique, various tRNA molecules, one of
which is radioactively labeled with carbon-14 (14C), are
mixed with ribosomes and synthetic trinu-cleotides that are bound to a filter.
The mixture of tRNAs is passed through the filter, and some bind and others
pass through. If the radioactive label is detected on the filter, then it is
known that the particular tRNA did bind. If the radioactive label is found in a
solution that flowed through the filter, then the tRNA did not bind. This
technique depends on the fact that aminoacyl-tRNAs bind strongly to ribosomes
in the presence of the correct trinucleotide. In this situation, the
trinucleotide plays the role of an mRNA codon. The possible trinucleotides are
synthesized by chemical methods, and binding assays are repeated with each type
of trinucleotide. For example, if the aminoacyl-tRNA for histidine binds to the
ribosome in the presence of the trinucleotide CAU, the sequence CAU is
established as a codon for histidine. About 50 of the 64 codons were identified
by this method.
Codon–Anticodon Pairing and Wobble
A codon forms base pairs with a complementary anticodon of a tRNA when an amino acid is incorporated during protein synthesis. Because there are 64 possible codons, one might expect to find 64 types of tRNA but, in fact, the number is less than 64 in all cells.
If there are 64 codons, how can there be less than 64 tRNA molecules?
Some
tRNAs bond to one codon exclusively, but many of them can recognize more than
one codon because of variations in the allowed pattern of hydrogen bonding.
This variation is called “wobble”
(Figure 12.4), and it applies to the first base of an anticodon, the one at the
5' end, but not to the second or the third base. Recall that mRNA is read from
the 5' to the 3' end. The first (wobble) base of the anticodon hydrogen-bonds
to the third base of the codon, the one at the 3' end. The base in the wobble
position of the anticodon can base-pair with several different bases in the
codon, not just the base specified by Watson–Crick base pairing (Table 12.2).
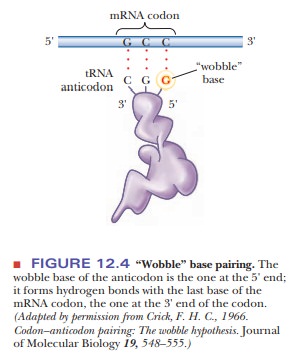
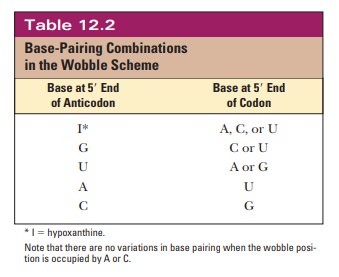
When the
wobble base of the anticodon is uracil, it can base-pair not only with adenine,
as expected, but also with guanine, the other purine base. When the wobble base
is guanine, it can base-pair with cytosine, as expected, and also with uracil,
the other pyrimidine base. The purine base hypoxanthine fre-quently occurs in
the wobble position in many tRNAs, and it can base-pair with adenine, cytosine,
and uracil in the codon (Figure 12.5). Adenine and cyto-sine do not form any
base pairs other than the expected ones with uracil and guanine, respectively
(Table 12.2). To summarize, when the wobble position is occupied by I (from
inosine, the nucleoside made up of ribose and hypoxan-thine), G, or U,
variations in hydrogen bonding are allowed; when the wobble position is
occupied by A or C, these variations do not occur.
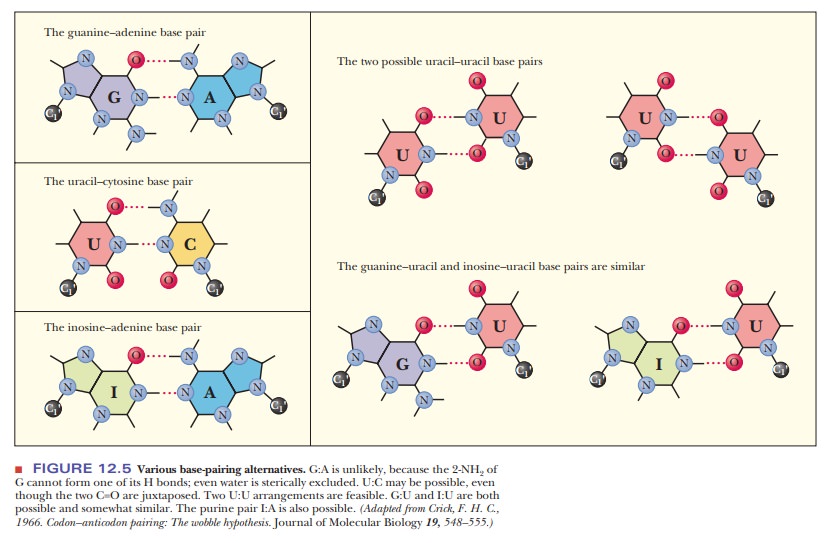
The
wobble model provides insight into some aspects of the degeneracy of the code.
In many cases, the degenerate codons for a given amino acid differ in the third
base, the one that pairs with the wobble base of the anticodon. Fewer different
tRNAs are needed because a given tRNA can base-pair with several codons. As a
result, a cell would have to invest less energy in the synthesis of needed
tRNAs. The existence of wobble also minimizes the damage that can be caused by
misreading of the code. If, for example, a leucine codon, CUU, were to be
misread as CUC, CUA, or CUG during transcription of mRNA, this codon would
still be translated as leucine during protein synthesis; no damage to the
organism would occur. We saw in earlier that drastic consequences can result
from misreading the genetic code in other codon positions, but here we see that
such effects are not inevitable.
A universal code is one that is the same in all organisms. The universality of the code has been observed in viruses, prokaryotes, and eukaryotes. However, there are some exceptions. Some codons seen in mitochondria are different from those seen in the nucleus.
There are also at least 16 organisms that have code
variations. For example, the marine alga Acetabularia
translates the stan-dard stop codons, UAG and UAA, as a glycine rather than as
a stop. Fungi of the genus Candida
translate the codon CUG as a serine, where that codon would specify leucine in
most organisms. The evolutionary origin of these differences is not known at
this writing, but many researchers believe that understanding these code
variations is important to understanding evolution.
Summary
The genetic code is based on a series of three bases coding for an
amino acid.
The code is nearly universal in all organisms
from viruses through humans. The code has no punctuation, meaning the mRNA is
read three bases at a time with no spaces in between. The code is
nonoverlapping as well, meaning that each base is part of only one codon.
The genetic code was determined by a variety of
techniques, such as using synthetic mRNA with known sequences to see what
proteins would be translated from them.
Although
there are 64 combinations of three bases leading to 64 codons, there are fewer
types of tRNA anticodons. This means that standard Watson–Crick base pairing
must be broken on occasion. The wobble model of codon–anticodon base pairing
shows that some bases at the 5' end of the anticodon of the tRNA can base-pair
with multiple bases on the codon.
Related Topics