Chapter: Biotechnology Applying the Genetic Revolution: DNA, RNA, and Protein
Regulation of Transcription in Prokaryotes
REGULATION OF TRANSCRIPTION IN PROKARYOTES
In prokaryotes, a variety of activator and repressor proteins control which genes get transcribed. The activators and repressors all work by binding to DNA in the promoter region and either stimulating or blocking the action of bacterial RNA polymerase. In E. coli, about 1000 of the 4000 total genes are expressed at one time. Activator proteins work by positive regulation; in other words, genes are expressed only when the activator gives a positivesignal. In contrast, repressors work by negative regulation. Here the gene is expressed only when the repressor is removed. Some repressors block RNA polymerase from binding to the DNA; others prevent initiation of transcription even though RNA polymerase has bound.
Regulation of transcription is complex, even in simple prokaryotes. Many genes are controlled by a variety of factors. Some operons in bacteria have multiple repressors and activators. Less often, regulatory proteins may block elongation either by slowing the actual rate of elongation or by signaling premature termination. Conversely, a few antiterminator proteins are known that override termination and allow genes downstream of the termination site to be expressed.
Prokaryotes use both positive regulation, where activator proteins signal RNA polymerase to transcribe the gene, or negative regulation, where the transcription factor inhibits RNA polymerase.
Prokaryotic Sigma Factors Regulate Gene Expression
Prokaryotic RNA polymerase has the sigma subunit, which recognizes the promoter first and binds the catalytic portion of the enzyme (the core enzyme). There are many different sigma subunits, and each one recognizes a different set of genes. The σ 70 subunit, or RpoD, is the most commonly used form. It recognizes most of the housekeeping genes in E. coli. During the stationary phase, when E. coli is not growing rapidly, σ 38, or RpoS, activates the necessary genes. (Sigma subunits are named either by σ plus their molecular weight or by Rpo [for RNA polymerase] plus their function: D = default, S = stationary, etc.)
Another sigma factor, RpoH, or σ 32, activates genes needed during heat shock. Normally E. coli grows at body temperature, 37°C, and stops growing at temperatures much above 43°C. At such higher temperatures proteins begin to unfold and are degraded. RpoH activates expression of chaperoninsthat help proteins fold correctly and prevent aggregation. RpoH also activates proteases that degrade proteins too damaged by the heat to be saved.
The transcription and translation of RpoH depend on temperature. When E. coli grows at a normal temperature, very few misfolded proteins are present. DnaK (a chaperonin) and HflB (a protease) are found in the cytoplasm but have very few bad proteins to “fix”; therefore, they bind to RpoH and degrade it. They even degrade partially translated RpoH protein. When high temperatures promote unfolding and aggregation of proteins, DnaK and HflB bind to the aberrant proteins and no longer destroy RpoH. Now the sigma factor initiates transcription of other genes associated with heat shock.
Sigma (σ) subunits are transcription factors that associate with prokaryotic RNA polymerase and control which genes are transcribed.
Lactose Operon Demonstrates Specific and Global Activation
Many genes require specific regulator proteins to activate transcription via RNA polymerase. Some of these proteins exist in two forms: active (binds to DNA in promoter region) and inactive (nonbinding). The forms are interconverted by small signal molecules or inducers that alter the shape of the protein. For example, the inducer allo-lactose controls the lactose operon.
The lactose or lac operon is well characterized because it was the first to be studied. It has a promoter upstream of three structural genes, lacZ, lacY, and lacA. The lacZYA genes are transcribed as a polycistronic message. Upstream of the promoter is the gene for LacI protein, the lac operon repressor (Fig. 2.6). The lacZ gene encodes b-galactosidase, which cleaves the disaccharide lactose into galactose and glucose. The lacY gene encodes lactose permease, which transports lactose across the cytoplasmic membrane into the bacteria. Finally, the lacA gene encodes the protein lactose acetylase, with an unknown role. The promoter has a binding site, lacO, for the repressor protein, which overlaps the binding site for RNA polymerase. This region is also known as the operator, and when the repressor binds, RNA polymerase cannot transcribe the operon.
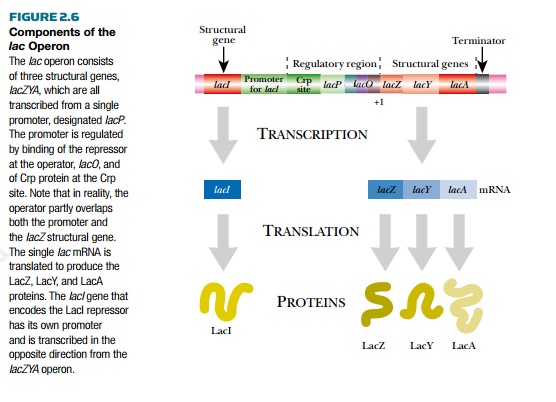
There is also a binding site for CRP protein (cyclic AMP receptor protein), also known as CAP (catabolite activator protein). This is a global regulator that activates transcription of many different operons for using alternate sugar sources. It is active when E. coli does not have glucose to utilize as an energy source.
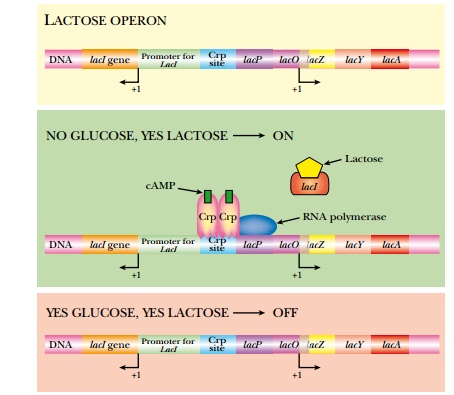
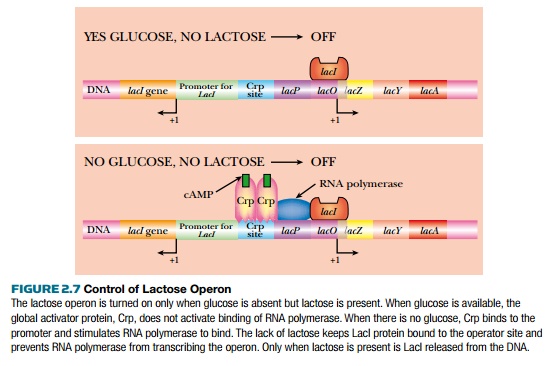
The environment controls whether or not the lactose operon is expressed (Fig. 2.7). When E. coli has plenty of glucose, then the lactose operon is turned off (as well as other operons for other sugars such as maltose or fructose). When glucose is present, levels of a small inducer, cyclic AMP (cAMP), are low. If E. coli runs out of glucose, the levels of cAMP increase, and bind to Crp, the global regulator. Crp, in turn, dimerizes so that it can bind to the Crp sites in various promoters, such as the lactose operon. Crp binding alone will not activate the lactose operon; lactose must also be present to activate transcription. If lactose is available, β-galactosidase catalyzes a side reaction, converting some lactose into allo-lactose. This acts as an inducer and binds to the tetrameric LacI repressor protein. This releases the repressor from the promoter. The lactose operon is expressed only when both glucose levels are low and lactose is present.
The control relies on two inducer molecules: cAMP binds to the global activator, Crp, and allo-lactose binds to the specific repressor, LacI. One control is global (Crp) because it controls many different operons, and one control is specific (LacI) because it only regulates the lactose operon.
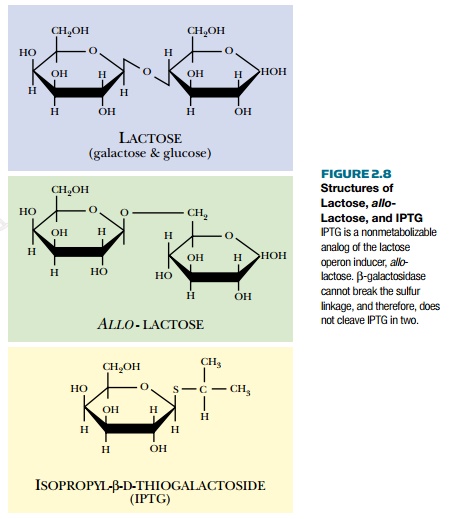
Many researchers use the lactose promoter to control expression of other genes. In the lab, allo-lactose is not used to induce the promoter. Agratuitous inducer, IPTG (isopropyl thiogalactoside), is used (Fig. 2.8). IPTG is not cleaved by β-galactosidase because its two halves are linked through a sulfur rather than oxygen. Since it is not metabolized, IPTG does not have to be added continually throughout the experiment (as would be the case for a natural inducer).
The lac operon is important to understand because its inducers and regulators are used to control new genes that are engineered into model organisms.
Control of Activators and Repressors
A variety of mechanisms controls gene activators and repressors. In some cases, the repressor or activator binds to the promoter of its own gene and controls its own transcription; this is called autogenous regulation.
Many activators and repressors rely on activation by small molecules, as for Crp and lacI. In some cases, a repressor needs a co-repressor in order to be active. For example, ArgR, represses the arginine biosynthetic operon when arginine is present. Arginine is a co-repressor and ensures that the bacteria do not make the amino acid when it is not needed.
In many cases, adding different groups, such as phosphate, methyl, acetyl, AMP-, and ADP-ribose, covalently modifies activators or repressors. Thetwo-component regulatory systems of bacteria transfer phosphate groups from a sensor protein to a regulator protein (Fig. 2.9). The first protein, thesensor kinase, senses a change in the environment and changes shape. This causes the kinase to phosphorylate itself using ATP. The phosphate group is then transferred to the regulator protein (an activator or repressor), which changes shape to its DNA binding form. The phosphorylated regulator then binds to its recognition site in the target promoter. This either stimulates or represses transcription of the operon.
There are many different two-component systems in bacteria that respond to a variety of environmental conditions.
For example, when there is low oxygen, the ArcAB system modifies gene expression to compensate. The ArcB protein is the sensor kinase, and it has three phosphorylation sites. The ArcA regulator has only one site for phosphorylation. The phosphate group is passed from one site to the next in aphosphorelay system, ultimately regulating transcription of the genes. These types of phosphorelays are very common, particularly in eukaryotes where there are often more than two components.
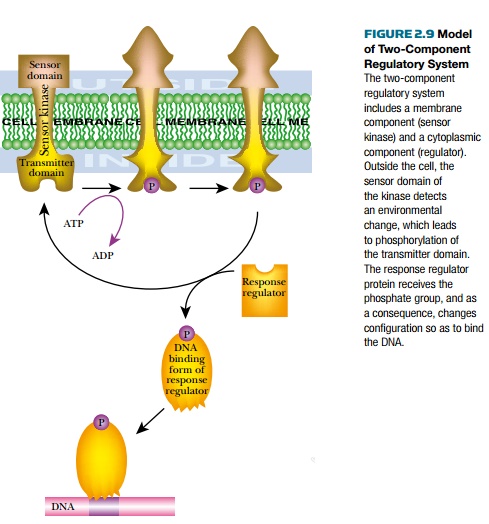
Prokaryotic regulators are controlled by different mechanisms so that the gene of interest is expressed only when necessary.
Related Topics