Chapter: Biotechnology Applying the Genetic Revolution: Environmental Biotechnology
Identifying New Genes with Metagenomics
IDENTIFYING
NEW GENES WITH METAGENOMICS
Metagenomics is the study of the genomes of whole communities of microscopic
life forms. Approaches include
shotgun DNA sequencing, PCR, RT-PCR, and other genetic methods. Metagenomic
research sometimes allows us to identify microorganisms, viruses, or free DNA
that exist in the natural environment by identifying genes or DNA sequences
from the organisms. Metagenomics applies the knowledge that all creatures
contain nucleic acids that encode various protein products; therefore,
organisms do not have to be cultured, but can be identified by a particular
gene sequence, protein, or metabolite. The term meta-, meaning more comprehensive, is also used in meta-analysis, which is the process of
statistically combining separate analyses. Metagenomics is the same as genomics
in its approach. The difference between genomics and metagenomics is the nature
of the sample.
Genomics focuses on one
organism, whereas metagenomics deals with a mixture of DNA from multiple
organisms, “gene creatures” (i.e., viruses, viroids, plasmids, etc.), and/or
free DNA. Most microorganisms have never been cultured or previously
identified. Using metagenomics, researchers investigate, catalogue, and analyze
the current microbial diversity. They identify new proteins, enzymes, and
biochemical pathways. They also hope to provide insight into the properties and
functions of the new organisms. The knowledge garnered from metagenomics has
the potential to affect how we use the environment to our benefit or harm.
Metagenomics has been used to
identify new beneficial genes from the environment, such as novel antibiotics,
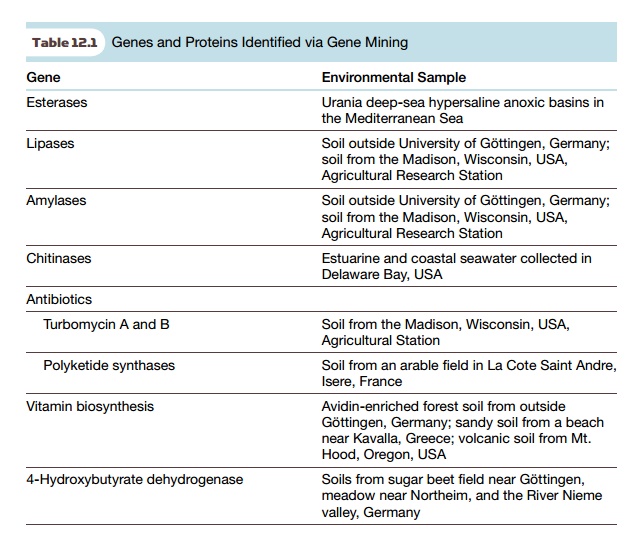
enzymes that biodegrade pollutants, and enzymes that make novel products (Table
12.1). Historically, studying microbes in the environment has identified many
useful products. In the early 1900s, Selman Waksman was studying actinomycetes
in soil when he discovered the antibiotic streptomycin. Similarly, metagenomics
research has identified (by accident) another antibiotic, called turbomycin.
The researchers were looking for hemolysin-related genes in the soil by
screening a metagenomic library (see later discussion). Hemolysin is a bacterial
toxin that punctures holes in susceptible cell membranes, allowing the cellular
contents to leak out and the cell then dies. Hemolysin lyses red blood cells
and creates a clear zone around a bacterial colony growing on blood agar
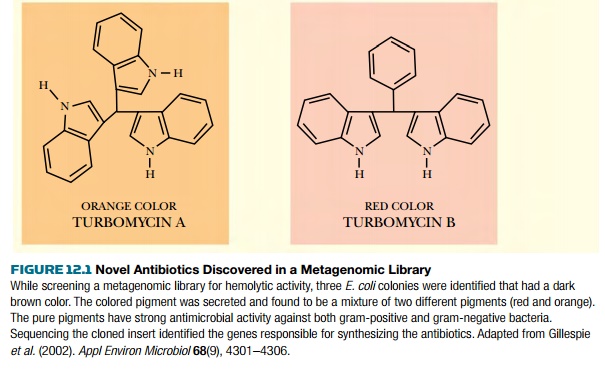
plates. Some E. coli clones from the
library had dark red or orange colors. Further investigation of these clones
found two novel antibiotics, turbomycin A and B (Fig. 12.1).
Novel biodegradation pathways
have also been found in bacteria that inhabit contaminated sites. Enzymes that
can reduce the toxic effects of oil- and petroleum-based contaminants are found
in bacteria that utilize the pollutants as an energy source. Even bacteria that
thrive in environments contaminated with radioactivity have been identified.

Related Topics