Chapter: Plant Biochemistry: Protein biosynthesis occurs in three different locations of a cell
Proteins are degraded by proteasomes in a strictly controlled manner
Proteins are degraded by proteasomes in a strictly controlled manner
In a eukaryotic cell the protein outfit is regulated not only by its synthesis but also by its degradation. Eukaryotes possess a highly conserved machinery for controlled protein degradation, consisting of a multienzyme complex termed proteasome. The outstanding role of this pathway in plants may be illustrated by the fact that more than 5% of all structural genes in Arabidopsis partici-pate in this degradation device. In order to be degraded, the corresponding proteins are labeled by covalent attachment of ubiquitin molecules. Ubiquitin occurs as a highly conserved protein in all eukaryotes. It has an identical sequence of 76 amino acids in all plants. The C-terminus of the molecule contains a glycine residue, which is the terminal carboxyl group exposed to the outside. Proteins destined for degradation are conjugated to ubiquitin by forming a so-called iso-peptide link between the glycine carboxyl group and the amino group of a lysine residue of the target protein.
The attachment of ubiquitin to a target protein requires the interplay of three different enzymes (Fig. 21.14A). The ubiquitin-activating enzyme (E1) activates ubiquitin upon the consumption of ATP to form a thioester with an SH-group of the enzyme. The ubiquitin is then transferred to a ubiquitin-conjugating enzyme (E2). Subsequently the target protein and the ubiquitin attached to E2 react with a specific ubiquitin-protein ligase (E3) to form the -isopeptide linkage bond. More uniquitin molecules can be con-jugated, either to lysine residues of the ubiquitin already attached to the target protein or by linkage to other lysine residues of the target protein. In this way target proteins can be labeled with a chain of ubiquitin molecules or by several ubiquitin molecules at various sites. Genome analyses indi-cated that Arabidopsis contains two genes for E1, 24 for E2, and 1,200 genes for E3. Apparently, the specificity of protein degradation is governed by the various E3 proteins.
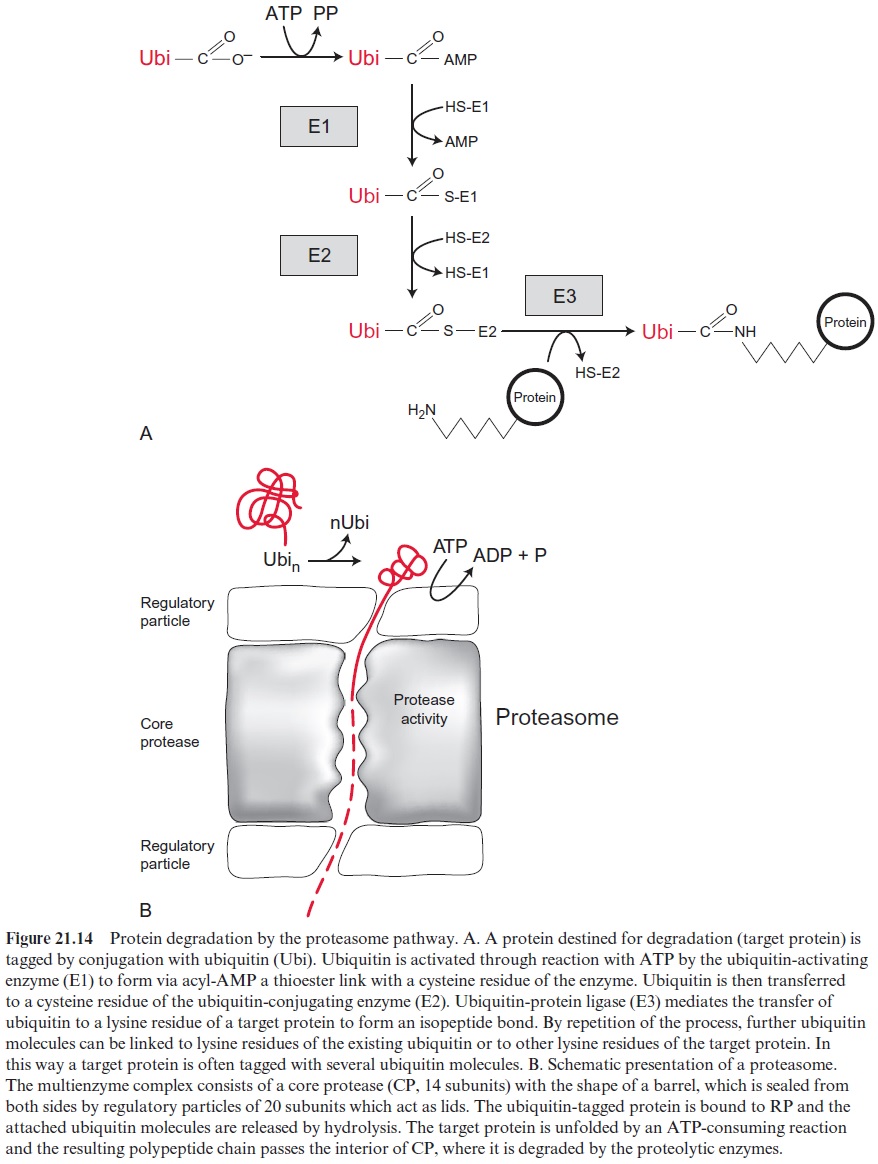
The proteolysis of the labeled target protein is catalyzed by the proteasome. This multienzyme complex can be divided into two different particles, a core protease (CP) consisting of 14 subunits and a regulatory particle (RP) consisting of 20 subunits. The core protease has a barrel-like structure, with the catalytic sites for proteolysis inside it. The openings on both sides are sealed by a regulatory particle (Fig. 21.14B). The regula-tory particle recognizes the ubiquitin-labeled target proteins, and catalyzes the hydrolytic cleavage of the ubiquitin molecules, which are thus avail-able for further ubiquitination of proteins. The target protein bound to the RP subunit is unfolded at the expense of ATP, and the peptide chain is allowed to pass through the interior of the barrel, where it is split by the proteolytic activity into peptides of 7 to 9 amino acid residues, which are released from the barrel and further digested by cytosolic peptidases. In organelles proteins are also subjected to a quality control: proteins that carry a defect or are not used anymore are degraded upon the consump-tion of ATP by a machinery resembling the cytosolic proteasome.
The controlled protein degradation by the proteasome plays a role in the regulatory functions of the phytohormones gibberellin andauxin. These hormones induce via signal transduction chains the degradation of tran-scription repressors and in this way enhance the expression of genes . Proteasomes are also involved in the degradation of activated phytochrome A.
The ubiquitin-dependent proteasome pathway is not the only way to degrade cellular proteins. During senescence, proteins of the cytoplasm or organelles are surrounded by membranes to form autophagic vesicles, which fuse to lytic vacuoles, wherein the proteins are subsequently degraded by proteolysis.
Related Topics