Chapter: Plant Biochemistry: Protein biosynthesis occurs in three different locations of a cell
Protein synthesis is catalyzed by ribosomes
Protein synthesis is catalyzed by ribosomes
Ribosomes are large riboprotein complexes that consist of three to four different rRNA molecules and a large number of proteins. In the intervals between the end of the translation of one mRNA and the start of the trans-lation of another mRNA, the ribosomes dissociate into two subunits. The ribosomes of the cytosol, plastids, and mitochondria are different in size and composition (Table 21.1). The cytosolicribosomes (termed eukaryotic ribosomes), with a sedimentation constant of 80S, dissociate into a small subunit of 40S and a large subunit of 60S. In contrast, the mitochondrial ribosomes, with a size of about 78S, varying from species to species, and the plastidic ribosomes (70S) are smaller. Due to their relationship to the bac-terial ribosomes, the mitochondrial and plastidic ribosomes are classified asprokaryotic ribosomes. Ribosomes of the bacteria, e.g., Escherichia coli, have a sedimentation constant of 70S.
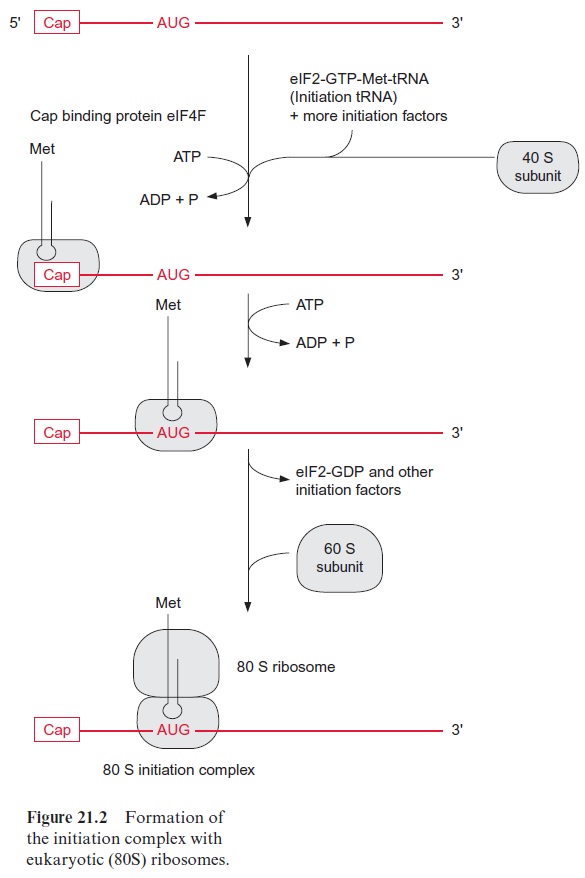
At the beginning of translation, the mRNA forms an initiation complex with a ribosome. A number of initiation factors participate in this process. The eukaryotic translation occurring in the cytosol will be described first. At the beginning of the process, the eukaryotic initiation factor2 (eIF2), together with GTP and a transfer RNA loaded with methionine, form an initiation-transfer-RNA-complex. With the participation of other initiation factors, it is then bound to the small 40S subunit of the ribosome. The initiation factor eIF4F, which consists of several protein components (also known as Cap-binding protein), mediates the binding of the initiation-tRNA-complex to the Cap sequence of the 5 end of the mRNA (see Fig. 20.9). For this the consumption of ATP is required. Driven by the hydrolysis of another ATP, the 40S subunit migrates downstream (5’ → 3’ ) until it finds the AUG start codon. Usually the first AUG triplet on the mRNA is the start codon, but in some mRNAs, the translation starts at a later AUG triplet. The neighboring sequences on the mRNA decide which AUG triplet is recognized as the start codon. The large 60S subunit is then bound to the 40S subunit, accompanied by the dissociation of several initiation factors and GDP. The formation of the initiation complex is now completed and the resulting ribosome is able to translate (Fig. 21.2).
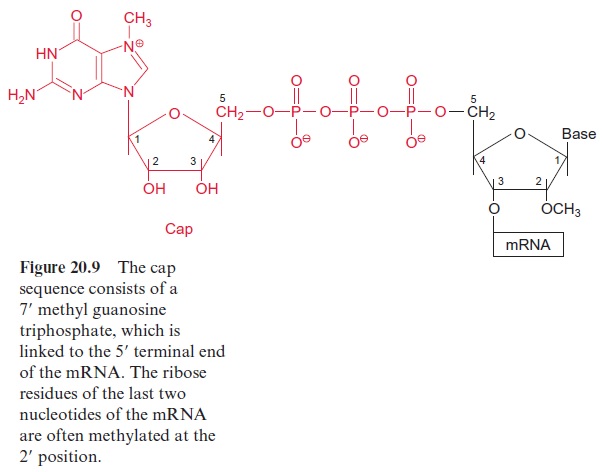
The mitochondrial and plastidic mRNAs have no cap sequence. Plastidic mRNAs have a special ribosome binding site for the initial binding to the small subunit of the ribosome consisting of a purine-rich sequence of about 10 bases. This sequence, called the Shine-Dalgarno sequence, binds to the 16S-rRNA of the small ribosome subunit. A Shine-Dalgarno sequence is also found in bacterial mRNAs, but it is not known whether it also plays this role in the mitochondria. In mitochondria, plastids and bacteria, the initiation tRNA is loaded with N-formyl methionine (instead of methionine as in the cytosol). After peptide formation the formyl residue is cleaved from the methionine.
A peptide chain is synthesized
A ribosome, completed by the initiation process, contains two sites where the tRNAs can bind to the mRNA. The peptidyl site (P) allows the binding of the initiation tRNA to the AUG start codon (Fig. 21.3). The aminoacyl (A) site covers the second codon of the gene. On the other side of site P is the exit (E) site where the empty tRNA is released. The elongation begins after the corresponding aminoacyl-tRNA occupies the A site by form-ing base pairs with the second codon. Two elongation factors participate in this. The eukaryotic elongation factor (eEF1α ) binds GTP and guides the corresponding aminoacyl-tRNA to the A site, during which the GTP is hydrolyzed to GDP and P. The cleavage of the energy-rich anhydride bond in GTP enables the aminoacyl-tRNA to bind to the codon at the A site. Afterwards the GDP, still bound to eEF1α , is exchanged for GTP, as mediated by the elongation factor eEF1βγ . The eEF1α -GTP is now ready for the next cycle.
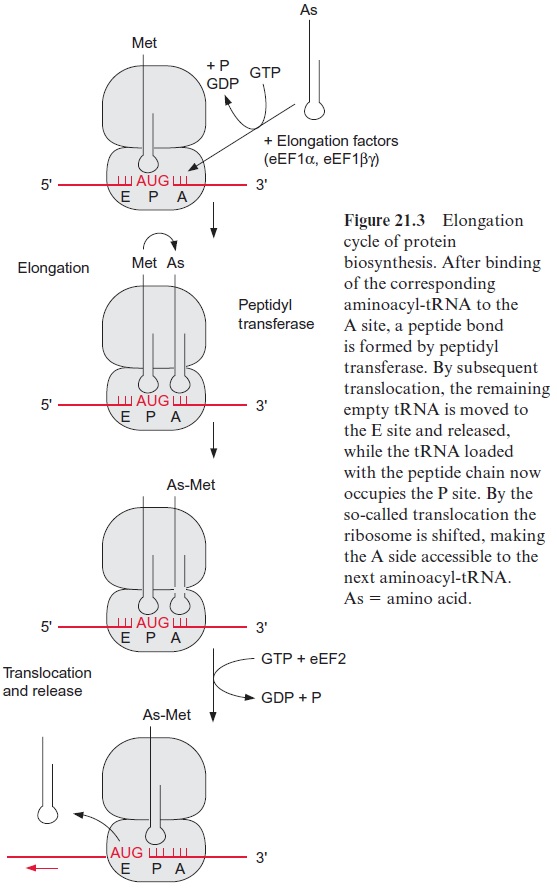
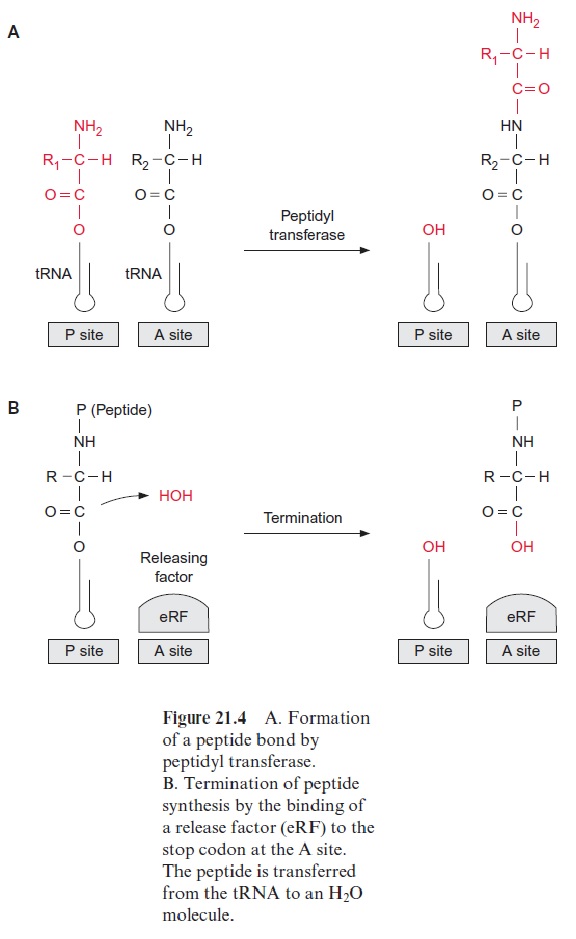
Subsequently a peptide linkage is formed between the carboxyl group of methionine and the amino group of the amino acid of the tRNA bound to the A site. The peptidyl transferase, which, as part of the ribosome, cata-lyzes this reaction, is a complex enzyme consisting of several ribosomal pro-teins. The 25S-rRNA has a decisive function in the catalysis. The enzyme facilitates the N-nucleophilic attack on the carboxyl group, whereby the peptide bond is formed. This results in the formation of a dipeptide bound to the tRNA at the A site (Fig. 21.4A).
Accompanied by the hydrolysis of one molecule of GTP into GDP and P, the elongation factor eEF2 facilitates the translocation of the ribosome along the mRNA to three nucleotides downstream (Fig. 21.3). In this way the free tRNA arrives at site E, is released, and the tRNA loaded with the dipeptide now occupies the P site. The third aminoacyl-tRNA binds to the vacant site A and a further elongation cycle can begin.
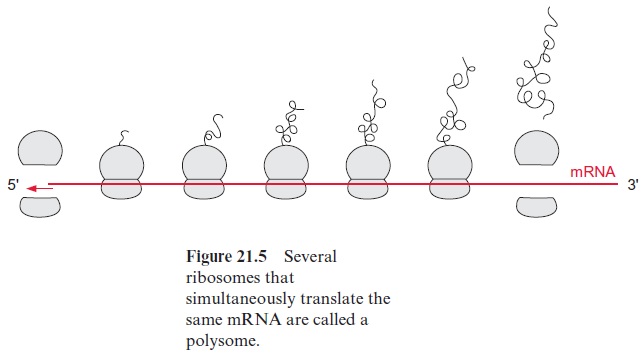
After several elongation cycles, the 5’ end of the mRNA is no longer bound to the ribosome and can start a new initiation complex. An mRNA that is translated simultaneously by several ribosomes is called apolysome (Fig. 21.5).
Translation is terminated when the A site finally reaches a stop codon (UGA, UAG, or UAA) (Fig. 21.4B). These stop codons bind the release factor (eRF) accompanied by hydrolysis of GTP to form GDP and P. Binding of eRF to the stop codon alters the specificity of the pepti-dyl transferase: a water molecule instead of an amino acid is now the acceptor for the peptide chain. In this way the protein formed is released from the tRNA.
Specific inhibitors of the translation can be used to decide whether a protein is encoded in the nucleus or the genome of plastids or mitochondria
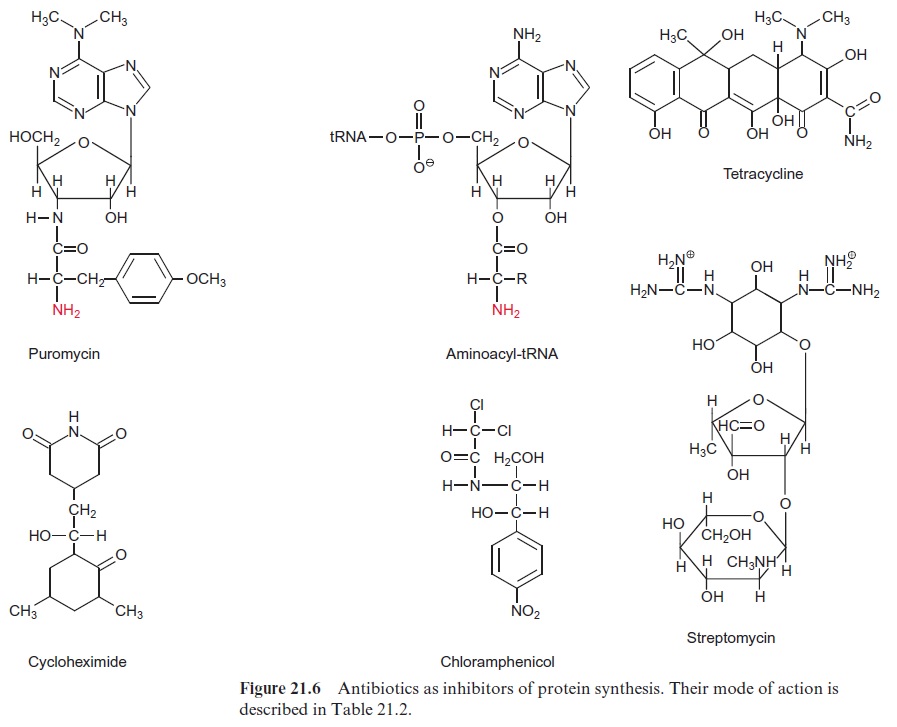
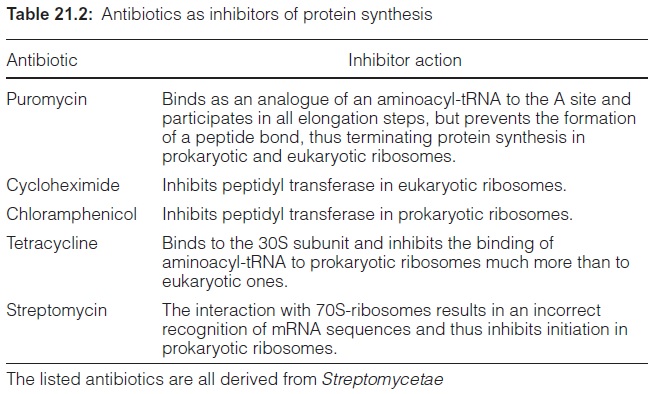
Elongation, translocation and termination occur in prokaryotic ribosomes in an analogous way as in eukaryotic ribosomes. Only minor alterations are observed, e.g., the termination of the prokaryotic ribosomes does not require GTP. Furthermore, eukaryotic and prokaryotic translation can react differ-ently with certain antibiotics (Fig. 21.6, Table 21.2). Puromycin, an analogue of tRNA, is a general inhibitor of protein synthesis, whereas cycloheximide inhibits only protein synthesis by eukaryotic ribosomes. Chloramphenicol, tet-racycline, and streptomycin primarily inhibit protein synthesis byprokaryotic ribosomes. These inhibitors can be used to determine whether a certain pro-tein is encoded in the nucleus or in the genome of plastids or mitochondria. A relatively simple method for monitoring protein synthesis is to measure the incorporation of a radioactively labeled amino acid (e.g., 35S-labeled methio-nine) into proteins. If the incorporation of this amino acid into a protein is inhibited by cycloheximide, this indicates that the protein is encoded in the nucleus. Likewise, inhibition by chloramphenicol shows that the correspond-ing protein is encoded in the genome of plastids or mitochondria.
The translation is regulated
The synthesis of many proteins is specifically regulated at the level of trans-lation. This regulation may involve protein kinases by which proteins par-ticipating in translation are phosphorylated. Since the rate-limiting step of translation is primarily the initiation, this step is especially suited for a regulation of translation. In animals, the initiation factor eIF2 is inacti-vated by phosphorylation and initiation is therefore inhibited. Little is known about the regulation of translation in plants.
Related Topics