Chapter: Plant Biochemistry: Protein biosynthesis occurs in three different locations of a cell
Nuclear encoded proteins are distributed throughout various cell compartments
Nuclear encoded proteins are distributed throughout various cell compartments
The ribosomes present in the cytosol also synthesize proteins destined for cell organelles, such as plastids, mitochondria, peroxisomes, and vacu-oles, as well as proteins to be secreted from the cell. To reach their cor-rect location, these proteins must be specifically transported across various membranes.
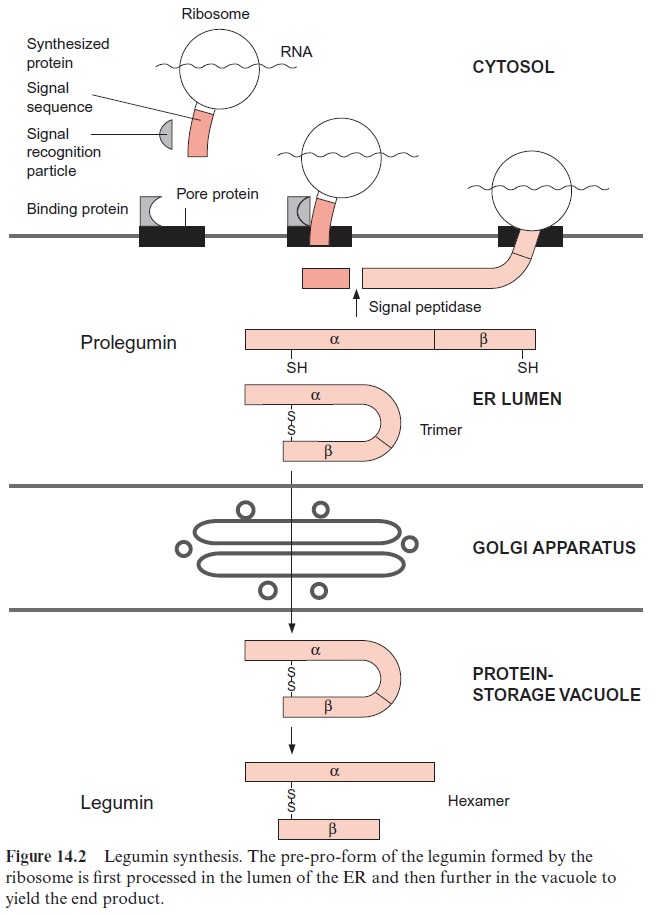
Proteins destined for the vacuole are transferred through the lumen of the ER . A signal sequence at the N-terminus of the newly synthesized protein binds specifically with a signal recognition particle and the whole complex to a pore protein (receptor) present in the ER membrane and thus directs the protein to the ER lumen. In such a case the ribosome is attached to the ER membrane (rough ER) during protein synthesis and the synthesized protein appears immediately in the ER lumen (Fig. 14.2). This process is called co-translational protein transport. These proteins are then transferred from the ER lumen by vesicle transfer across the Golgi appara-tus to the vacuole or are exported by secretory vesicles from the cell.
In contrast, protein uptake into plastids, mitochondria, and peroxi-somes occurs mainly, if not exclusively, by post-translational transport, which means that the proteins are transported across the membrane after completion of protein synthesis and their release from the ribosomes.
Most of the proteins imported into the mitochondria have to cross two membranes
More than 95% of the mitochondrial proteins in a plant are encoded in the nucleus and translated in the cytosol. Our present knowledge about the import of proteins from the cytosol into the mitochondria derives primarily from studies with yeast. In order to direct proteins from the cytosol to the mitochondria, they have to be provided with a mitochondrial presequence (transit peptide) as targeting signal. Some proteins destined for the mitochon-drial inner membrane or the inter-membrane compartment, as well as all the proteins for the mitochondrial outer membrane, contain internal targeting signals that have not yet been identified. Other proteins of the mitochondrial inner membrane and most of the proteins of the mitochondrial matrix are synthesized in the cytosol as precursor proteins, which contain 12 to 70 amino acids at their amino terminus as a transit peptide. These targeting prese-quences have a high content of positively charged amino acids and are able to form -helices in which one side is positively charged and the other side is hydrophobic. The three-dimensional structure of the amphiphilic -helices, rather than a certain amino acid sequence, functions as targeting signal. The directing function of this presequence can be demonstrated in an experiment. When a foreign protein, such as the dihydrofolate reductase from mouse, is provided with a targeting presequence for the mitochondrial matrix, this pro-tein is taken up into the mitochondrial matrix.
For the import of proteins into the mitochondrial matrix, both the outer and inner membranes have to be traversed (see Fig. 1.12). This pro-tein import occurs primarily at so-called translocation sites where the inner and outer membranes are closely attached to each other (Fig. 21.11). Each membrane provides its own translocation apparatus, which transfers the proteins in the unfolded state through the membranes.
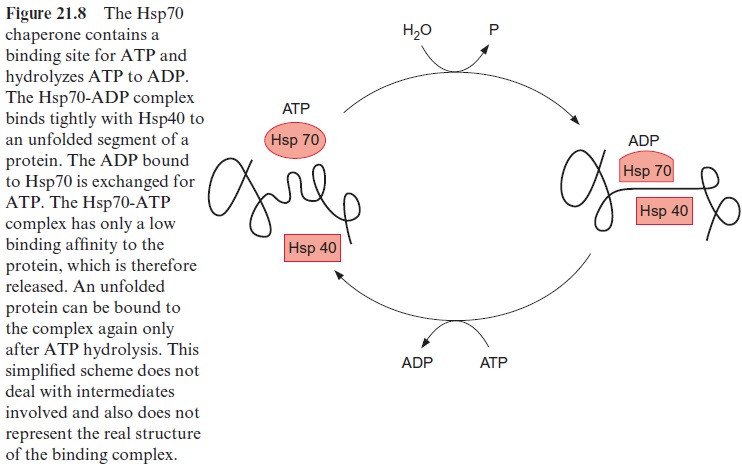
The precursor proteins synthesized by the ribosomes associate in the cytosol with chaperones (e.g., Hsp70) in order to prevent premature fold-ing or aggregation of the often hydrophobic precursor proteins. The asso-ciation with ctHsp70 is accompanied by the hydrolysis of ATP (Fig. 21.8). The transport across the outer membrane is catalyzed by a so-called TOM complex (translocase of the outer mitochondrial membrane) consisting of at least eight different proteins. The TOM20 and TOM22 subunits func-tion as receptors for the targeting presequence. An electrostatic interac-tion between the positively charged side of the α-helix of the presequence and the negative charge on the surface of TOM22 is probably involved in the specific recognition of the targeting signal. TOM22 and TOM20 then mediate the threading of the polypeptide chain into the translocation pore. Another receptor for the transport of proteins is TOM70. This receptor, together with TOM37, mediates the uptake of the ATP-ADP translocator protein and other translocators of the inner membrane, which contain an internal targeting signal instead of a presequence. Probably TOM40 as well as the small subunits TOM5, 6, 7 (not shown in Fig. 21.11) participate in the formation of the translocation pore.
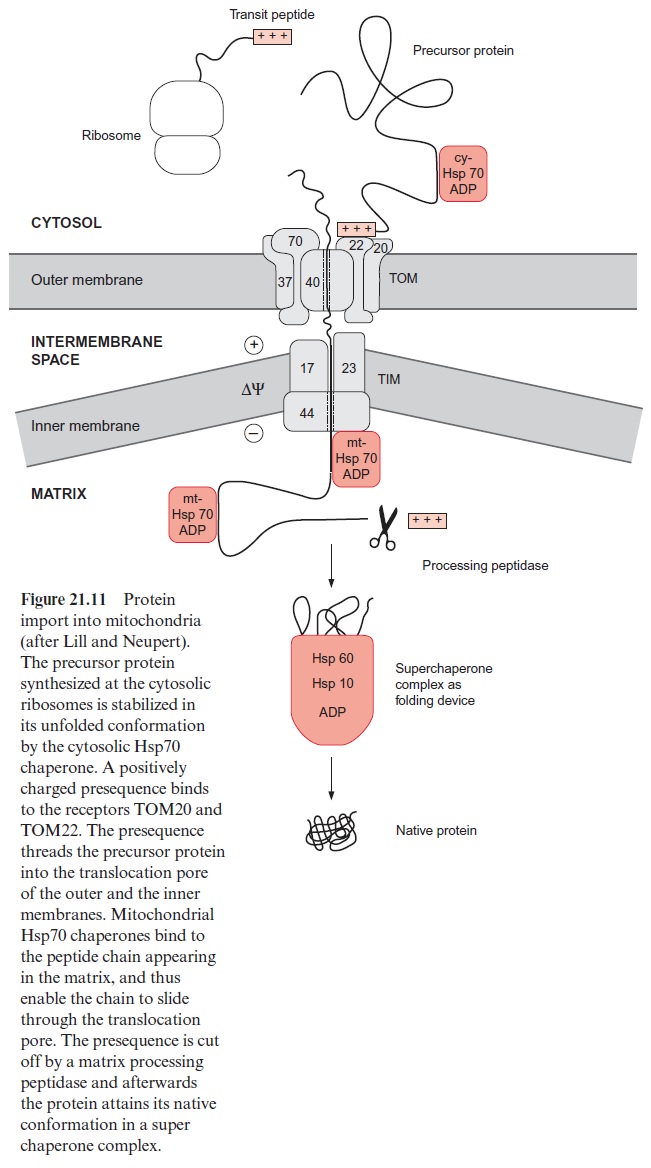
The subsequent transport across the inner membrane is catalyzed by the TIM complex (translocase of the inner mitochondrial membrane), consisting of the proteins TIM17, 23, 44 and several others not yet identified. A pre-condition for protein transport across the inner membrane is the presence of a membrane potential ΔΨ. Presumably the positively charged presequence is driven through the translocation pore by the negative charge at the matrix side of the inner membrane. The peptide chain appearing in the matrix is first bound to TIM 44 and is then bound with hydrolysis of ATP (Fig. 21.8) to an mtHsp70 chaperone and also to other chaperones not dealt with here. It is assumed that Brownian movement causes a section of the pep-tide chain to slip through the translocation pore, which is then immediately bound to the mtHsp70 inside, thus preventing the protein from slipping back. It is postulated that repetitive binding of Hsp70 converts a random move-ment of the protein chain in the translocation channel into a unidirectional motion. According to this model of a molecular ratchet, the ATP required for the reversible binding of mtHsp70 probably is not required for pulling the polypeptide chain through the pore, but, instead, to change its free diffusion across the two translocation pores into unidirectional transport. An alterna-tive hypothesis is also under discussion, according to which the protein enter-ing the pore is pulled into the matrix by ATP-dependent conformational changes of the mtHsp70 bound to the peptide.
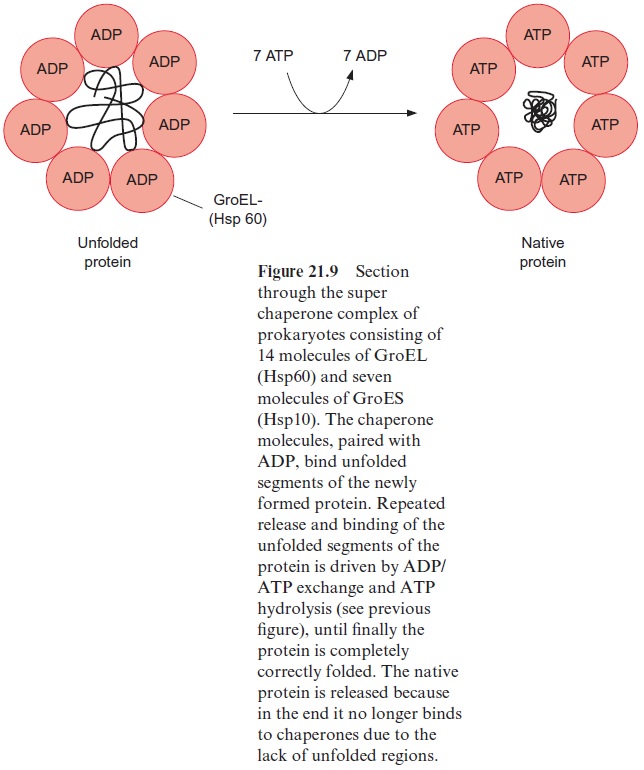
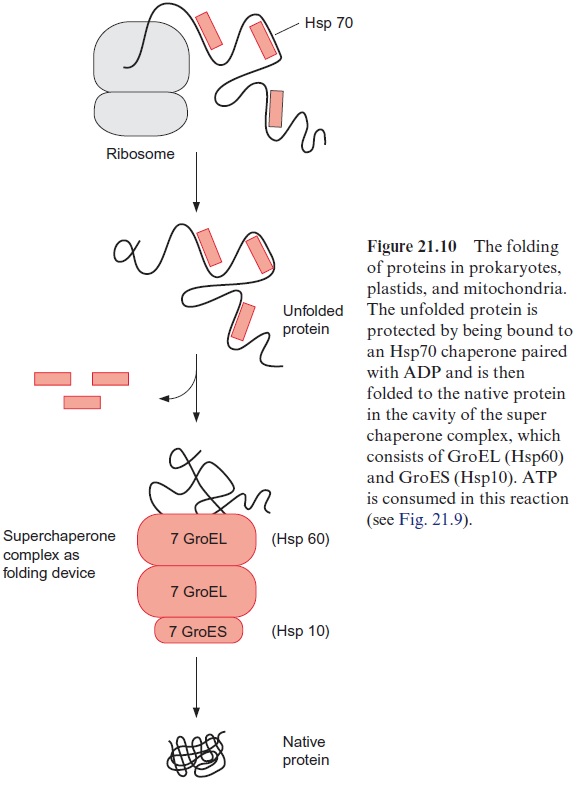
When the peptide chain arrives in the matrix, the transit peptide is imme-diately cleaved off from the protein by a processing peptidase (Fig. 21.11). The folding of the matrix protein probably occurs via a super chaperone folding apparatus consisting of the chaperones Hsp60 and Hsp10 (see Figs. 21.9 and 21.10). Proteins destined for the mitochondrial outer membrane, after being bound to the receptors of the TOM complex, are directly inserted into the membrane.
In most cases, proteins destined for the mitochondrial inner membrane, after transport through the outer membrane, are inserted directly from the inter membrane space into the inner membrane. In some cases, pro-teins destined for the inner membrane contain a presequence, which first directs them to the matrix space. After this presequence has been cut off by processing peptidases, they are then integrated from the matrix side into the inner membrane via a second targeting sequence.
The import of proteins into chloroplasts requires several translocation complexes
The transport of proteins into the chloroplasts shows parallels but also dif-ferences to the transport into mitochondria. Similar to mitochondria most chloroplast proteins are encoded in the nucleus and have to be transferred into the chloroplasts. Transport into the chloroplasts also proceeds post-translationally. The precursor proteins synthesized in the cytosol possess a targeting presequence, a transit peptide with 30 to 100 amino acid residues at the N-terminus of the protein. As in the mitochondria, the targeting signal probably does not consist of a specific amino acid sequence, but its function is due to the secondary structure of the presequence. The precursor proteins of the chloroplasts are stabilized by Hsp70 chaperones during their passage through the cytosol.
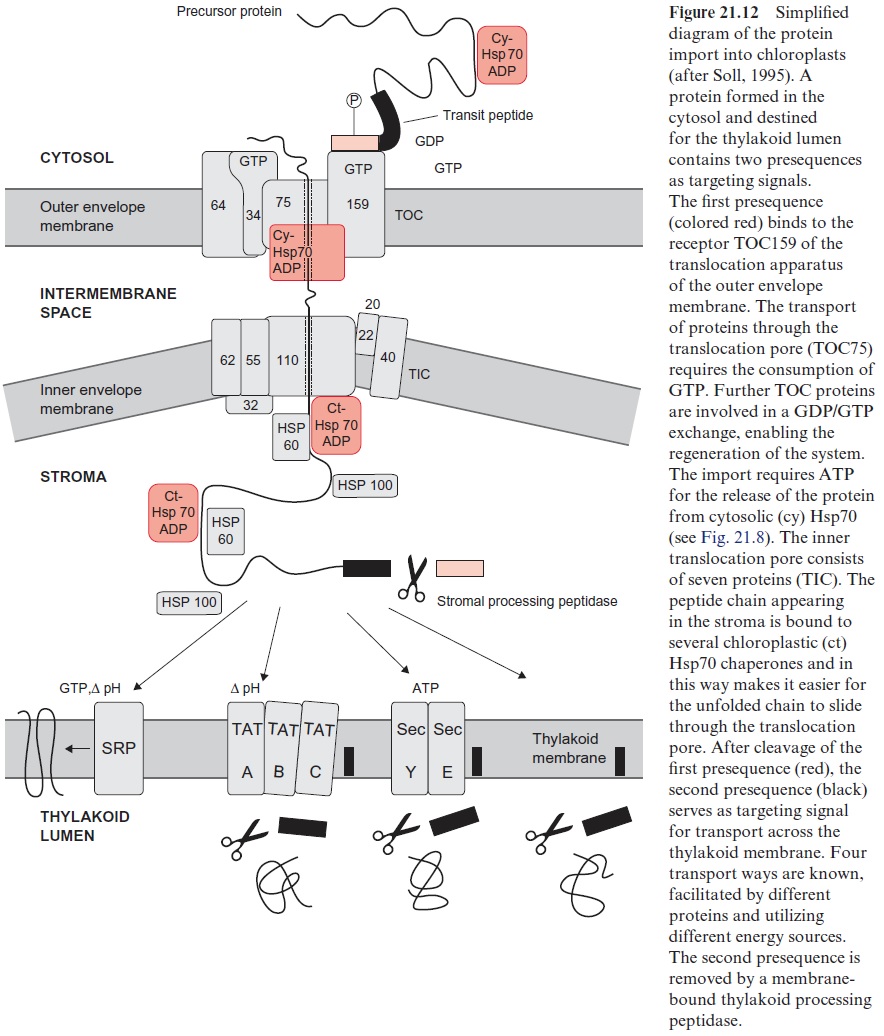
In order to be imported into the stroma, the protein must cross two membranes (Fig. 21.12). The translocation apparatus of the outer chlo-roplast envelope membrane contains at least 10 proteins, which, accord-ing to their molecular mass (in kDa), are named TOC (translocase of the outer chloroplast membrane) and together represent about 30% of the total membrane proteins of the outer envelope membrane. TOC175 functions as receptor and forms with TOC75 the translocation pore for the passage of the unfolded peptide chain. During the transport process further TOC proteins are involved, including those for binding and hydrolysis of GTP. Phosphorylated precursor proteins are recognized by a GTP-TOC34 com-plex and stimulate its GTPase activity. The change of free energy drives the protein from the resultant GDP-TOC34 complex across the outer mem-brane. In the intermembrane space the protein is bound to an HSP70. The subsequent transport across the inner envelope membrane involves at least seven TIC-(translocase of the inner chloroplast membrane) proteins, also named according to their molecular mass. In contrast to mitochondrial protein transport, protein transport into the chloroplast stroma does not require a membrane potentialΔΨ . Also in the chloroplasts, a unidirec-tional motion of the unfolded peptide chain through the translocation pore is caused by a repetitive binding of Hsp70 chaperones. According to the model of a molecular ratchet this process is accompanied by the hydrolysis of ATP. After delivering the protein chain to the stroma, the presequence is removed by a processing peptidase of the stroma. The resulting protein is folded to the native conformation, with the aid of an Hsp60-Hsp70-Hsp100 super chaperone complex, and is then released. In this way also the small subunit of RubisCO is delivered to the stroma, where it is assembled with the large subunit encoded in the chloroplasts.
Those proteins destined for the thylakoid membrane are first delivered to the stroma and then directed by four different mechanisms via internal targeting signals into the thylakoid membrane or lumen (Fig. 21.12). The SRP (secretion recognition particle) way inserts proteins (e.g., light har-vesting proteins) into the thylakoid membrane. This process, resembling the SRP dependent translocation of proteins into the ER , is driven by a pH gradient and requires GTP. The TAT (twin arginine trans-location) way transfers proteins into the lumen, facilitated by TAT pro-teins and driven by a pH gradient. Similar to this theSEC (secretion) way is facilitated by proteins that are similar to proteins of the secretion path-way and ATP is required. Some proteins are inserted into the thylakoid membrane spontaneously. In all ways except that of the SRP the presequence with the thylakoid addressing signal is cut off by peptidases.
Proteins are imported into peroxisomes in the folded state
The peroxisomes, in contrast to mitochondria and chloroplasts, contain no individual genome. All the peroxisomal proteins are nuclear-encoded. Peroxisomes, like mitochondria and chloroplasts, canmultiply by division and thus are inherited from mother cells. There are also observations that a de novo synthesis of peroxisomal membranes can take place at the endo-plasmatic reticulum. Signal sequences cause some peroxisomal membrane proteins to be incorporated into certain sections of the ER membrane, which are subsequently detached as vesicles regarded as pre-peroxisomes. It is postulated that these vesicles fuse to the peroxisomes already present or that they can form new peroxisomes by fusion.
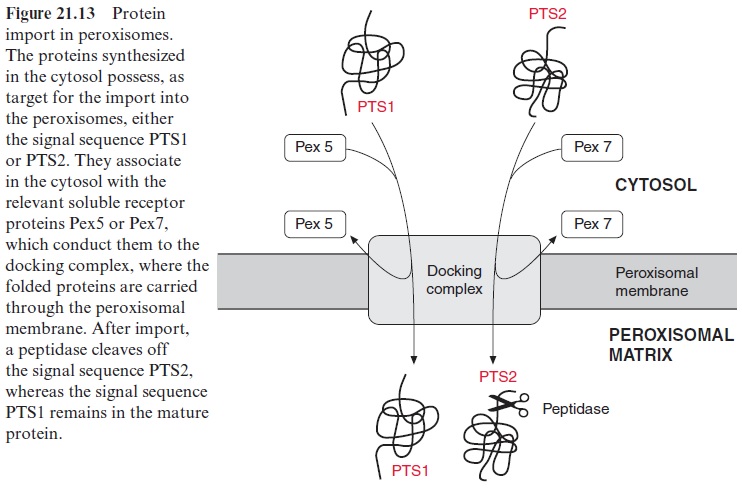
Independent of how the peroxisomes are formed, whether by divi-sion or by de novo synthesis, it is necessary to import the peroxisomal proteins, which are encoded in the nucleus and synthesized in the cytosol (Fig. 21.13). Two different signal sequences are known as targeting sig-nals (peroxisomal targeting signals) PTS1 and PTS2. PTS1 exhibits at the C-terminus the consensus sequence serine-lysine-leucine (SKL) which is not detached after the corresponding protein has been transported into the per-oxisomes. PTS2 consists of a sequence of about nine amino acids near the N-terminus of certain proteins and is removed after the import of the pro-tein via proteolysis. The proteins targeted by one of the two signals bind to the corresponding soluble receptor proteins (Pex5 and Pex7 peroxisomal biogenesis factor), which facilitate the binding to the translocation appara-tus (docking complex). The docking complex itself consists of several mem-brane proteins. After dissociation from the receptor proteins, the proteins are transferred upon the consumption of ATP across the membrane into the peroxisomal matrix, in a process not yet fully elucidated. According to present knowledge, the import of proteins into the peroxisomes proceeds in the folded state of the proteins, which is in contrast to the import into mito-chondria and chloroplasts where protein transport occurs in the unfolded conformation. It seems that protein import into the peroxisomes is entirely different from protein transport into the ER, mitochondria, and plastids.
Related Topics