Chapter: Plant Biochemistry: Nitrogen fixation enables plants to use the nitrogen of the air for growth
Legumes form a symbiosis with nodule-inducing bacteria
Legumes form a symbiosis with nodule-inducing bacteria
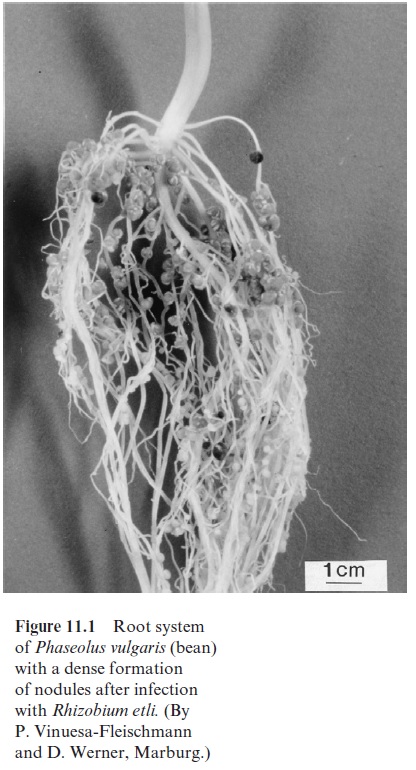
Initially it was thought that the nodules of legumes (Fig. 11.1) were caused by a plant disease, until their function in N2 fixation was recognized by Hermann Hellriegel (Germany) in 1888. He found that beans containing these nodules were able to grow without nitrogen fertilizer.
The nodule-inducing bacteria include, among other genera, Rhizobium, Bradyrhizobium, and Azorhizobium and are collectively called rhizobia. Species of Rhizobiumform nodules with peas, species of Bradyrhizobium with soybean and species of Azorhizobium with the tropical legume Sesbania. The rhizobia are strictly aerobic gram-negative rods, which live in the soil and grow heterotrophically in the presence of organic compounds. Some species (Bradyrhizobium) are also able to growautotrophically in the presence of H2, although at a low growth rate.
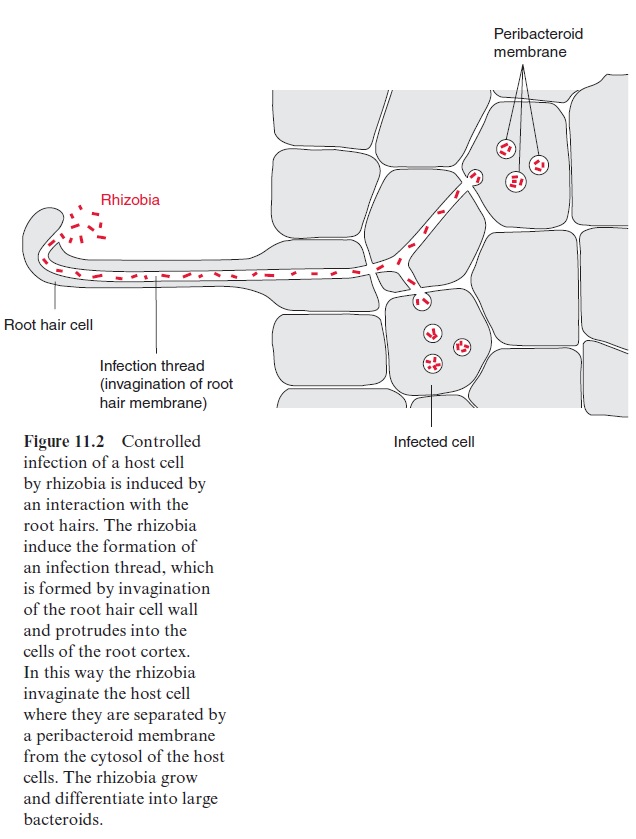
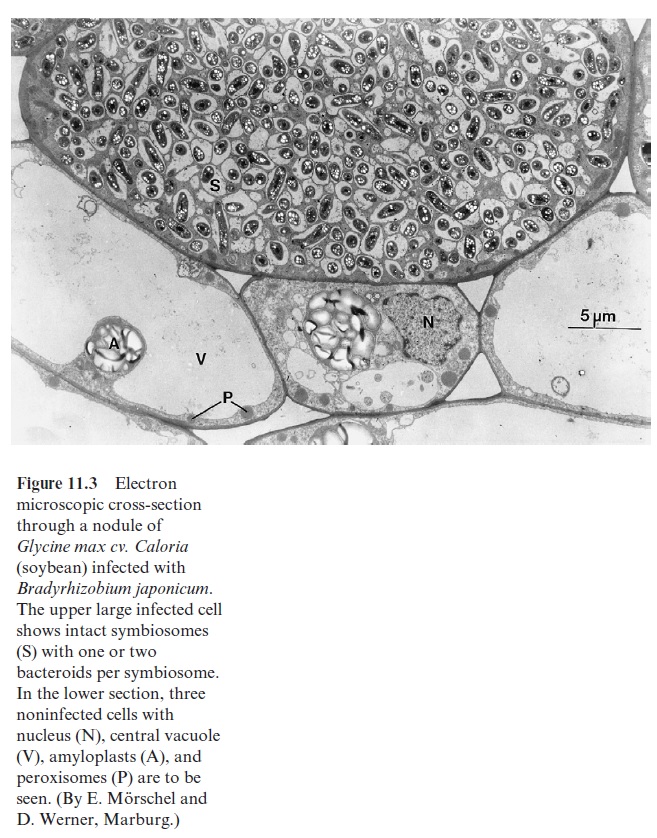
The uptake of rhizobia into the host plant is a controlled infection. The molecular basis of specificity and recognition is still only partially known. The rhizobia form species-specific nodulation factors (Nod factors). These are lipochito-oligosaccharides that acquire a high structural specificity (e.g., by acylation, acetylation, and sulfatation). They are like a security key with many notches and open the house of the specific host with which the rhizobia associate. The Nod factors bind to specificreceptor kinases of the host, which are part of signal transduction chains . In this way the “key” induces the root hair of the host to curl and the root cortex cells to divide, forming the nodule primordium. After the root hair has been invaded by the rhizobia, an infection thread forms (Fig. 11.2), which extends into the cortex of the roots, branches there and infects the cells of the nodule primordium. A nodule thus develops from the infection thread. The mor-phogenesis of the nodule is of similarly high complexity as any other plant organ such as the root or shoot. The nodules are connected with the root via vascular tissues, which supply them with substrates produced by pho-tosynthesis. The bacteria incorporated into the plant cell are enclosed by a peribacteroid membrane (also called a symbiosome membrane), which derives from the plasma membrane of the infected plant cell. The incorpo-rated bacteria are thus separated from the cytoplasm of the host cell in a so-called symbiosome (Fig. 11.3). In the symbiosome, the rhizobia differ-entiate to bacteroids. The volume of these bacteroids can be 10 times the volume of an individual bacterium. Several of these bacteroids are sur-rounded by a peribacteroid membrane.
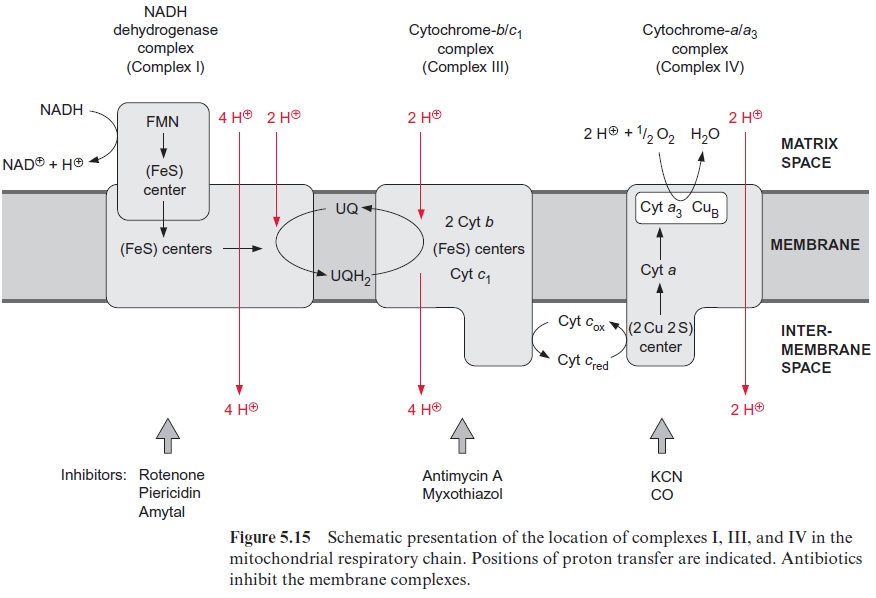
Rhizobia possess a respiratory chain which corresponds to the mito-chondrial respiratory chain (see Fig. 5.15). In a Bradyrhizobium species, an additional electron transport path develops during differentiation of the rhizobia to bacteroids. This path branches at the cyt-bc1 complex of the res-piratory chain and conducts electrons to another terminal oxidase, enabling an increased respiratory rate. It is encoded by symbiosis-specific genes.
The nodule formation relies on a balanced interplay of bacterial and plant gene expression
Symbiotic rhizobia provide a large number of genes, which are switched off in the free-living bacteria and are activated only after an interaction with the host, to contribute to the formation of an N2-fixing nodule. The bacte-rial genes encoding proteins required for N2 fixation are named nif and fix genes, and those that induce the formation of the nodules are called nod, nol and noe genes.
The host plant signals its readiness to form nodules by excreting several flavonoids as signal compounds for the chemo-attraction of rhizobia. These flavonoids bind to a bacterial nod gene protein, which is constitutively expressed (expressed at all times). The protein, to which the flavonoid is bound, activates the transcription of the othernod, nol and noe genes. The proteins encoded by these nod genes are involved in the synthe-sis of the Nod factors. Four, so-called “general” nod genes are present in nearly all rhizobia. In addition, more than 20 other nod genes are known, which are responsible for the host’s specificity.
Those proteins, which are required especially for the formation of nod-ules, and which are synthesized by the host plant in the course of nodule formation, are callednodulins. These nodulins include leghemoglobin (sec-tion 11.2), the enzymes of carbohydrate degradation (including sucrose synthase ), enzymes of the citrate cycle and the synthesis of glutamine and asparagines, and, if applicable, also of ureide synthesis. They also include an aquaporin of the peribacteroid membrane. Together, these proteins belong to the normal outfit of root cells, but are synthesized at elevated levels during nodule formation. The plant genes encoding these proteins are called nodulin genes. One differentiates between “early” and “late” nodulins. “Early” nodulins are involved in the process of infection and formation of nodules, and the expression of the corresponding genes is induced in part by signal compounds released from the rhizobia. “Late” nodulins are only synthesized after the formation of the nodules.
Metabolic products are exchanged between bacteroids and host cells
The main substrate provided by the host cells to the bacteroids is malate (Fig. 11.4), synthesized from sucrose, which is delivered by the sieve tubes.
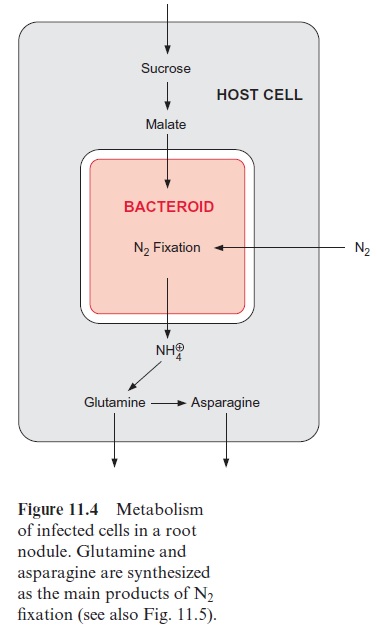
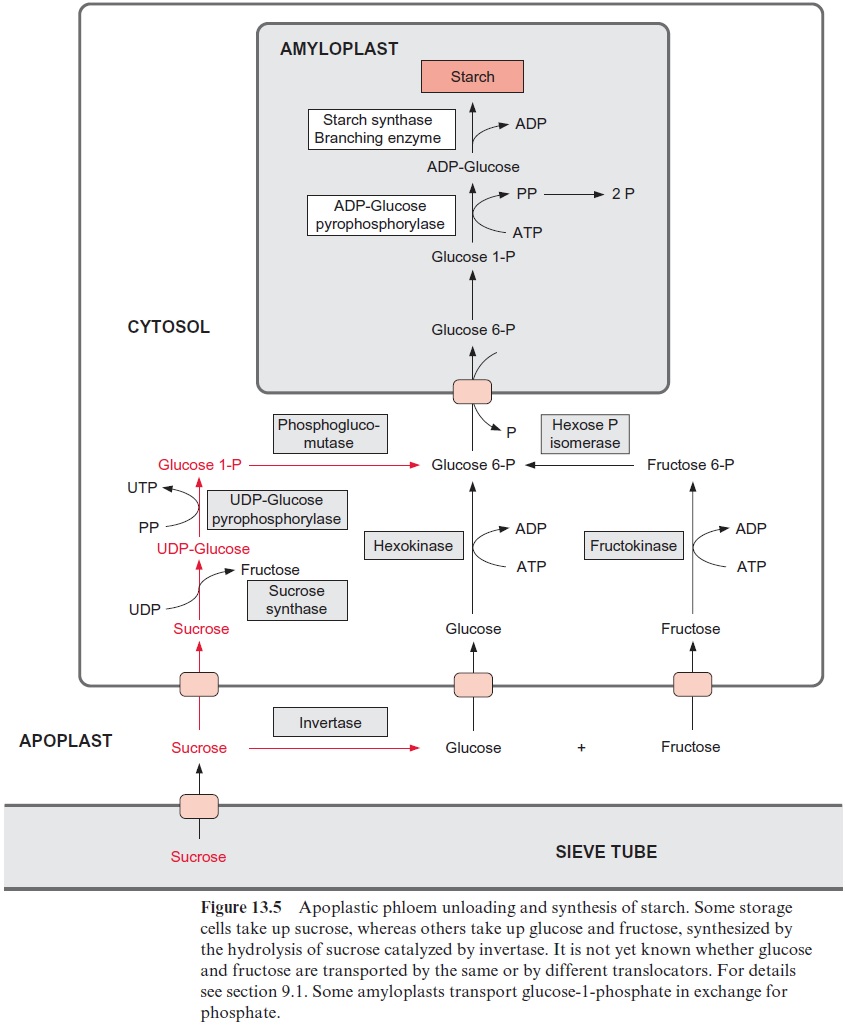
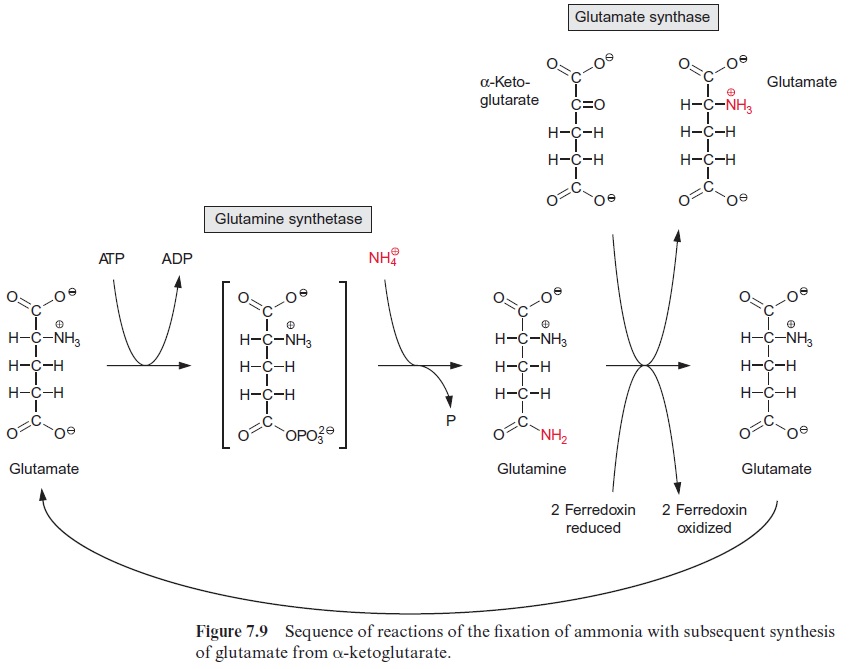
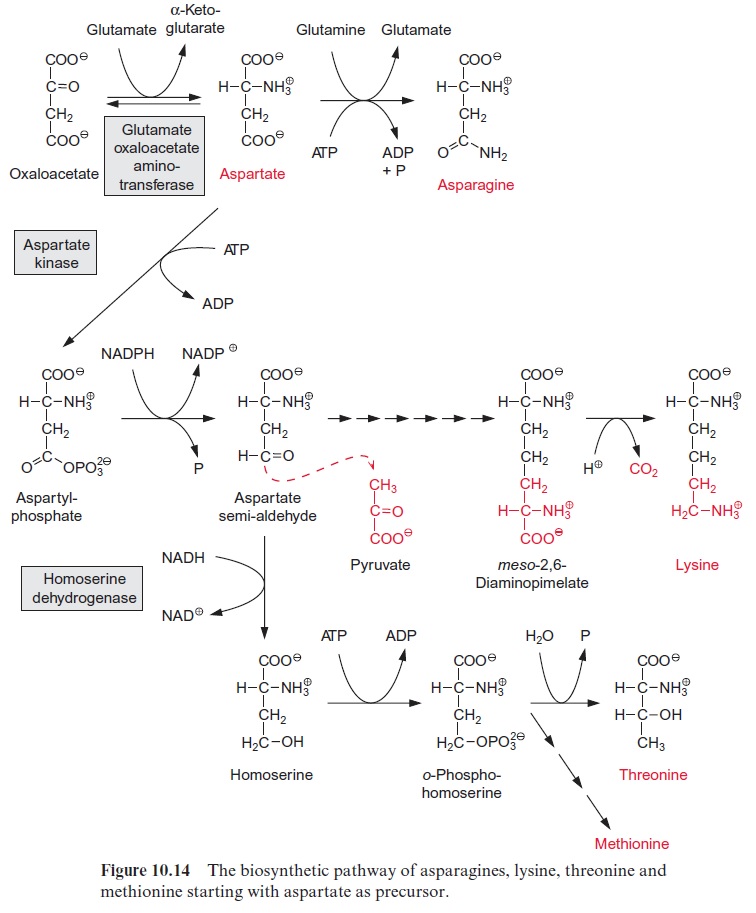
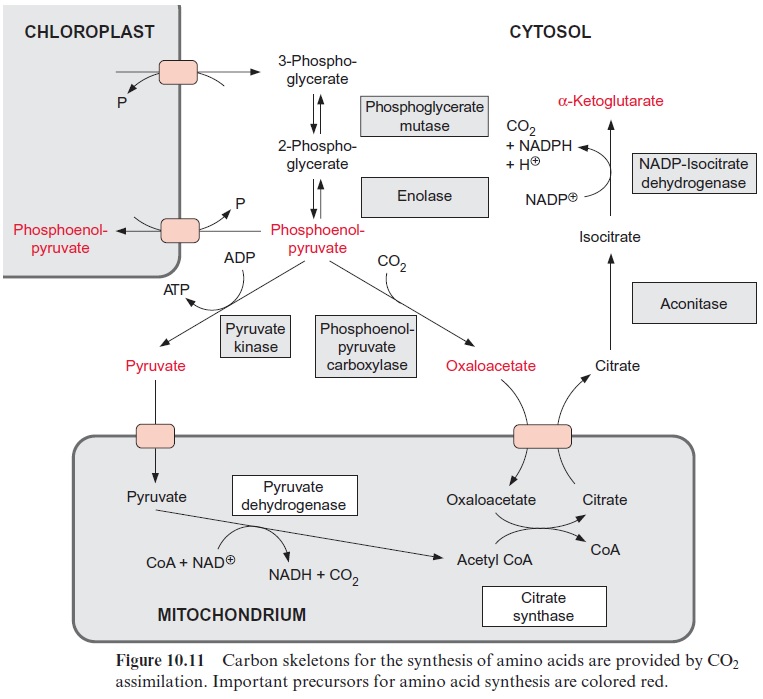
The sucrose is metabolized by sucrose synthase in the plant cell (Fig. 13.5), and further converted by glycolysis to phosphoenolpyruvate, which is sub-sequently carboxylated to oxaloacetate (see Fig. 10.11), and the latter is reduced to malate. Nodule cells contain high activities of phosphoenolpyru-vate carboxylase. NH4+ is delivered as a product of N2 fixation via a specific transporter to the host cell, where it is subsequently converted mainly into glutamine (Fig. 7.9) and asparagine (Fig. 10.14) and then transported via the xylem vessels to the other parts of the plant. It was recently shown that alanine can also be exported from bacteroids.
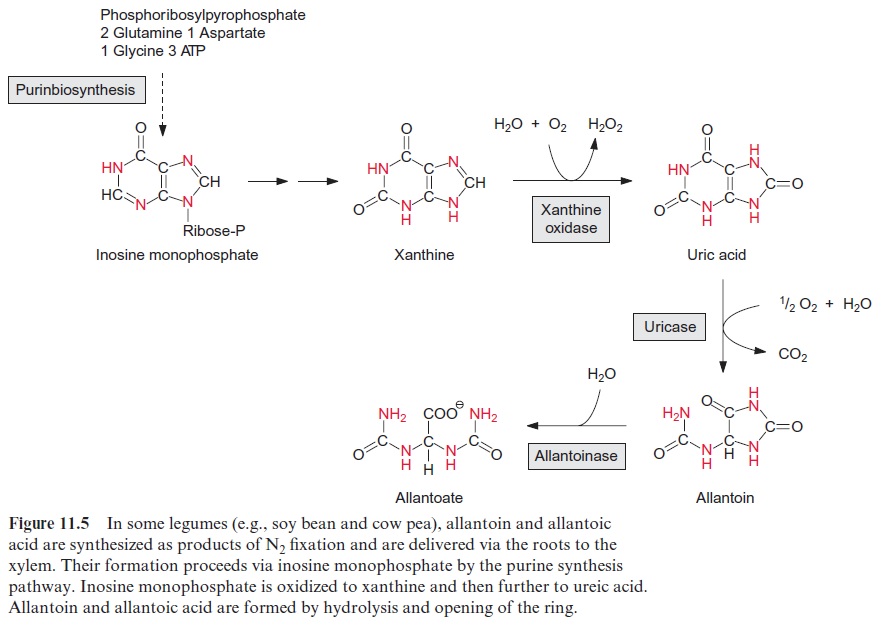
The nodules of some plants (e.g., soybean) export the fixed nitrogen as ureides (urea degradation products), especially allantoin and allantoic acid (Fig. 11.5). These compounds have a particularly high nitrogen to carbon (N/C) ratio. The formation of ureides in the host cells requires a compli-cated synthetic pathway. First, inosine monophosphate is synthesized via the pathway of purine synthesis, which is present in all cells for the synthe-sis of AMP and GMP, and then it is degraded via xanthine and ureic acid to the ureides.
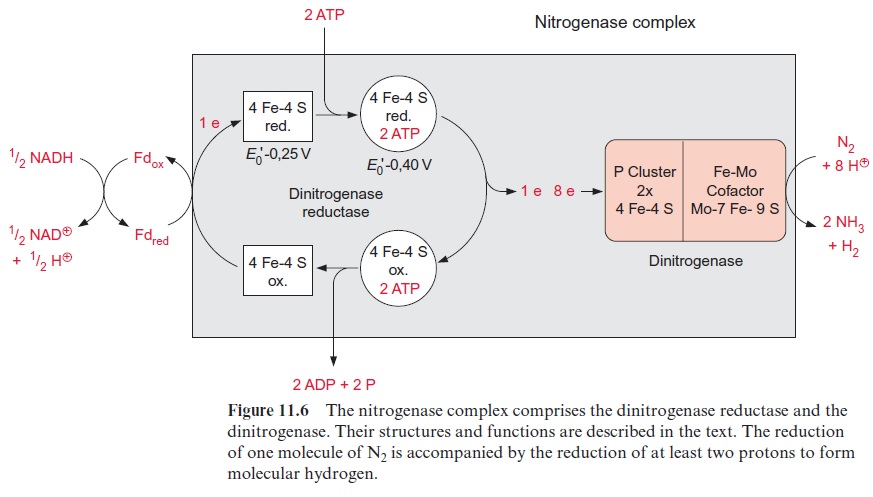
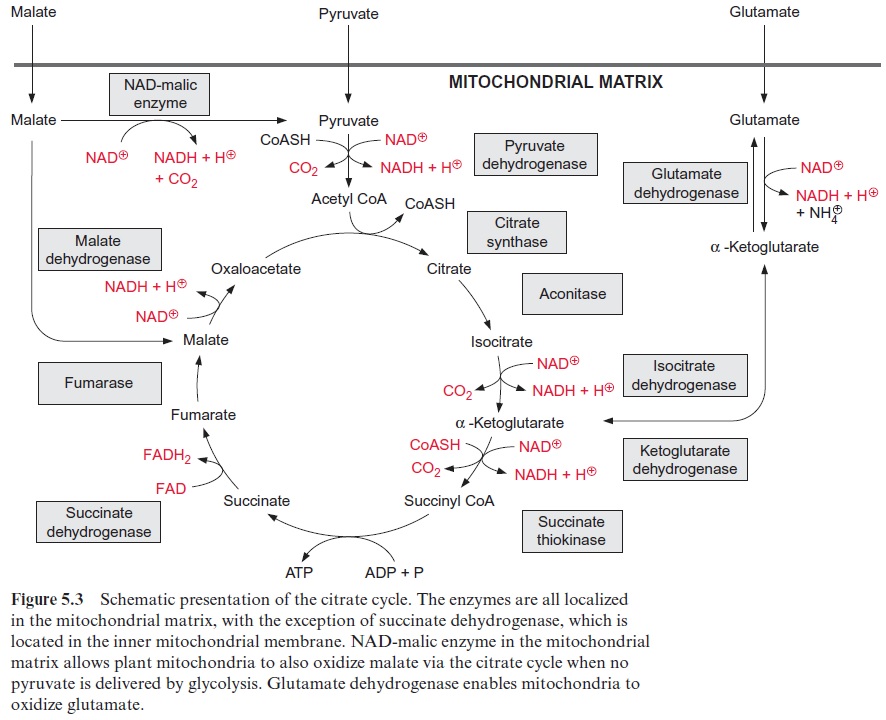
Malate taken up into the bacteroids (Fig. 11.4) is oxidized by the citrate cycle (Fig. 5.3). The reducing equivalents thus generated are the fuel for the fixation of N2 (Fig. 11.6).
Dinitrogenase reductase delivers electrons for the dinitrogenase reaction
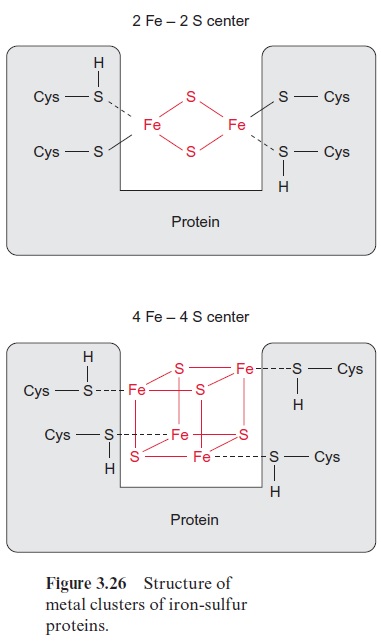
Nitrogen fixation is catalyzed by the nitrogenase complex, a very intricate system with nitrogenase reductase and dinitrogenase as the main compo-nents (Fig. 11.6). This complex is highly conserved and is present in the cytoplasm of the bacteroids. NADH formed in the citrate cycle delivers electrons via soluble ferredoxin to dinitrogenase reductase. The latter is a one-electron carrier, consisting of two identical subunits, which together form a 4Fe-4S cluster (see Fig. 3.26) and comprise two binding sites for ATP. After the reduction of dinitrogenase reductase, two molecules of ATP are bound, resulting in a conformational change of the protein, by which the redox potential of the 4Fe-4S cluster is raised from -0.25 to -0.40 V. After transfer of an electron to the dinitrogenase, the two ATP molecules are hydrolyzed to ADP and phosphate, and then released from the protein.
As a result, the conformation with the lower redox potential is restored and the enzyme is once more ready to take up one electron from ferredoxin. Thus, with the hydrolysis of two molecules of ATP, one electron is trans-ferred from NADH to dinitrogenase via dinitrogenase reductase.
N2 as well as H+ are reduced by dinitrogenase
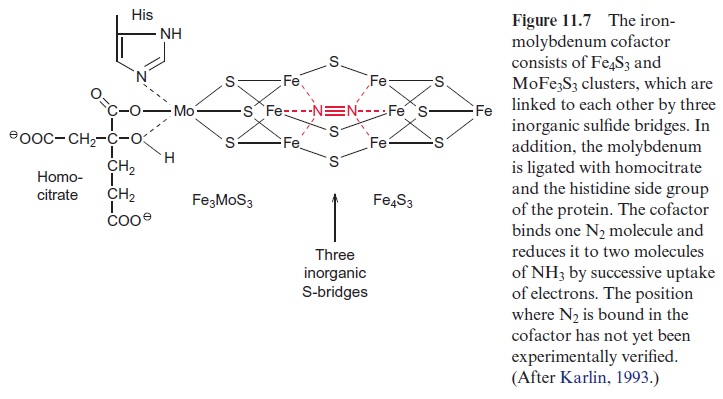
Dinitrogenase is an α2 β2 tetramer. The α and β subunits are similar in size and structure. The tetramer contains two catalytic centers, probably react-ing independently of each other, and each contains a so-called P clus-ter, consisting of two 4Fe-4S clusters and an iron molybdenum cofactor (FeMoCo). FeMoCo is a large redox center built of Fe4S3 and Fe3MoS3, which are linked to each other via three inorganic sulfide bridges (Fig. 11.7). Another constituent of the cofactor is homocitrate, which is linkedvia oxygen atoms of the hydroxyl and carboxyl group to molybdenum. Another ligand of molybdenum is the imidazole ring of a histidine residue of the protein. The function of the Mo atom is still unclear. Alternative nitrogenases are known in which molybdenum is replaced by vanadium or iron, but these nitrogenases are much more unstable than the nitroge-nase containing FeMoCo. The Mo atom possibly causes a more favora-ble geometry and electron structure of the center. It is not yet known hownitrogen reacts with the iron-molybdenum cofactor. One possibility would be that the N2 molecule is bound in the cavity of the FeMoCo center (Fig. 11.7) and that the electrons required for N2 fixation are transferred by the P cluster to the FeMoCo center.
The nitrogenase complex is able to reduce other substrates beside N2 (e.g., protons, which are reduced to molecular hydrogen):

During N2 fixation at least one molecule of hydrogen is formed per N2 reduced:

Thus the balance of N2 fixation is at least:
N2 + 4 NADH + 4 H+ + 16 ATP → 2 NH3 + H2 + 4 NAD+ + 16 ADP + 16 P
In the presence of sufficient concentrations of acetylene, only this is reduced and ethylene is formed:
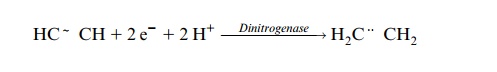
This reaction is used to measure the activity of dinitrogenase.
It is not known why H2 evolves during N2 fixation. It may be part of the catalytic mechanism or a side reaction or a reaction to protect the active center against the inhibitory effect of oxygen. The formation of molecular hydrogen during N2 fixation can be observed in a clover field. Many bacter-oids, however, possess hydrogenases by which H2 is reoxidized by electron transport:

It is questionable, however, whether in the bacteroids this reaction is coupled to the generation of ATP.
Related Topics