Chapter: Medical Immunology: Genetics of Immunoglobulins
Immunoglobulin Gene Rearrangement
IMMUNOGLOBULIN GENE REARRANGEMENT
It is well established that an immunoglobulin polypeptide chain is coded by multiple genes scattered along a chromosome of the germline genome. These widely separated gene seg-ments are brought together (recombined) during B-lymphocyte differentiation to form a complete immunoglobulin gene.
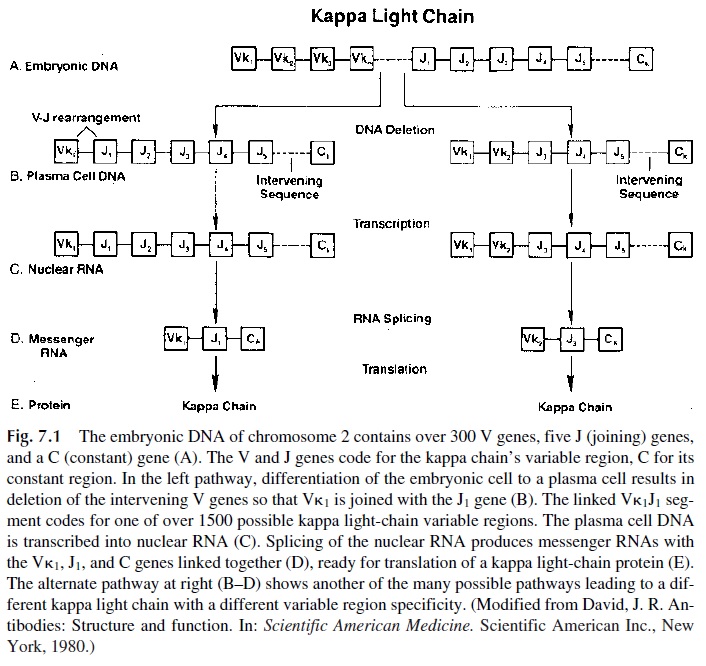
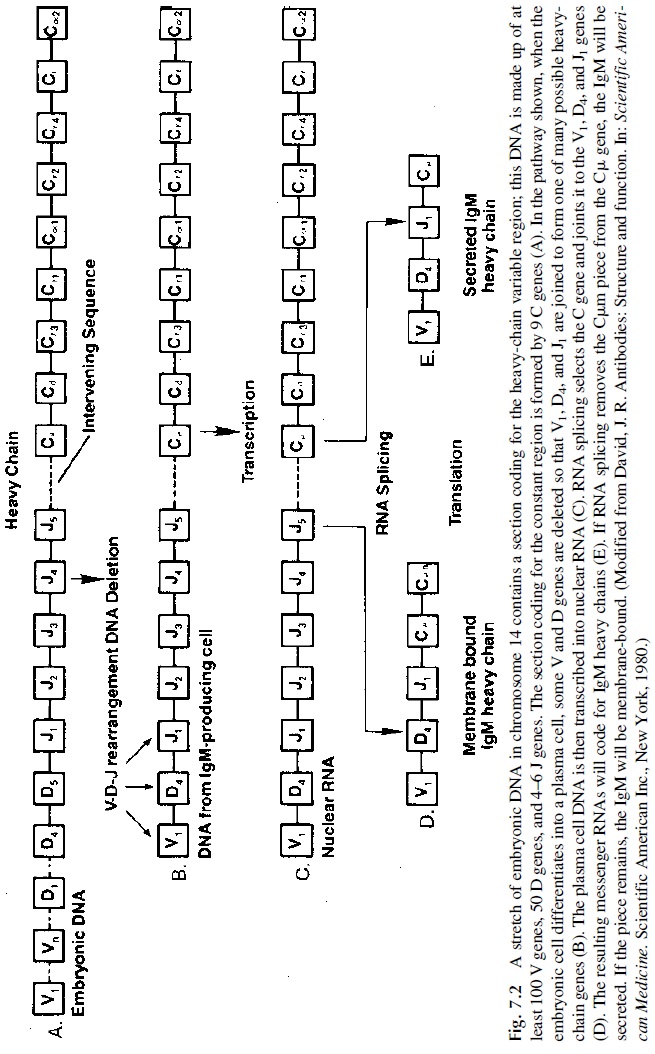
The V regions of the immunoglobulin light chains are coded by two gene segments, designated as V and J (J for joining, because it joins V and C region genes). Three gene segments are required for the synthesis of the V region of the heavy chains: V, J, and D (D for diversity, corresponds to the most diverse region of the H chain). To form a functional light- or heavy-chain gene, one or two gene rearrangements are needed. On chromosome 2 (where the k chain genes are located) or 22 (where the λ chain genes are located), a V gene moves next to a J gene. On chromosome 14 (where the heavy-chain genes are located), first the D and J regions are joined, and next one of the V genes is joined to the DJ complex. The VJ or VDJ segments, one of the C-gene complexes (Ck or Cl on chromosomes 2 or 22), and Cμ , Cδ , Cγ 3, Cγ 1, Cα 1, Cγ 2, Cγ 4, Ce , or Cα2 on chromosome 14 are then transcribed into nuclear RNA containing VJ- or VDJ-coding sequences, the interconnecting noncoding sequences, and the coding sequence of one of the C genes. The intervening noncoding se-quences are then excised making a contiguous VJC mRNA for an L chain and a contigu-ous VDJC mRNA for an H chain (Figs. 7.1 and 7.2). Gene rearrangements occur in a se-quential order: usually heavy chain genes rearrange first, followed by k chain genes and lastly by l genes.
It has been shown that the VDJ joining is regulated by two proteins encoded by two closely linked recombination-activating genes, RAG-1 and RAG-2, localized on the short arm of human chromosome 11.
These genes have at least two unusual characteristics not shared by most eukaryotic genes: they are devoid of introns and, although adjacent in lo-cation and synergistic in function, they have no sequence homology. The latter implies that, unlike the immunoglobulin and MHC genes, RAG-1 and RAG-2 did not arise by gene du-plication. Recent studies suggest that these genes may be evolutionarily related to trans-posons, genetic elements that can be transposed in the genome from one location to the other. Conserved recombination signal sequences (RSS) serve as substrate for the enzymes coded by the RAG genes. These enzymes introduce a break between the RSS and the cod-ing sequence. Mechanisms involved in subsequent rejoining to form a mature coding seg-ment are not completely understood.
The transcription of Ig genes, like other eukaryotic genes, is regulated by promoters and enhancers. Promoters, located 5’ of the V segment, are necessary for transcription ini-tiation. Enhancers, located in the introns between J and C segments, increase the rate of transcription. For this reason, immunoglobulin synthesis (H or L chains) is only detected after the VDJ or VJ rearrangements, which bring the promoter in close proximity to the en-hancer.
During ontogeny and functional differentiation, the H chain genes may undergo fur-ther gene rearrangements that result in immunoglobulin class switching. As the B lympho-cytes differentiate into plasma cells, one heavy-chain C-gene segment can be substituted for another without alteration of the VDJ combination (Fig. 7.3).
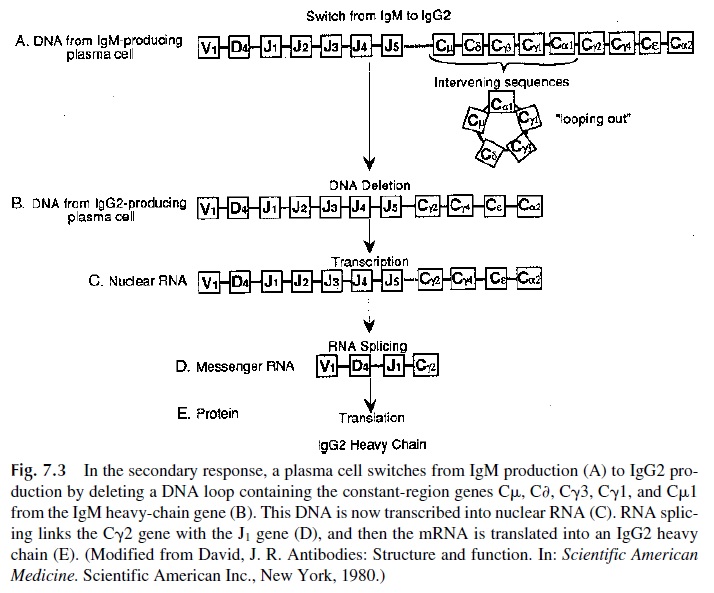
In other words, a given variable region gene can be expressed in association with more than one heavy-chain class or subclass, so that at the cellular level the same antibody specificity can be associated with the synthesis of an IgM immunoglobulin (characteristic of the early stages of ontogeny and of the primary response) or with an IgG immunoglobulin (characteristic of the mature in-dividual and of the secondary response). Three mechanisms have been proposed to explain the heavy-chain class switch: intrachromosomal DNA recombination (recombination be-tween sister chromatids, which constitute a chromosome), interchromosomal recombina-tion (recombination between nonsister chromatids of the two homologous chromosomes), and looping-out and deletion of intervening DNA sequences within a single chromosome. The mechanism that contributes most substantially to isotype switching is probably a loop-ing-out and deletion mechanism, in which the switched Ig sequences, Cμ and other inter-vening CH genes, loop out and are subsequently deleted. This mechanism would predict the formation of circular DNA molecules containing the sequences deleted by isotype switch-ing. Such DNA circles have now been isolated.
Related Topics